当前位置:
X-MOL 学术
›
Anal. Methods
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rapid identification and quantification of the antibiotic susceptibility of lactic acid bacteria using surface enhanced Raman spectroscopy†
Analytical Methods ( IF 2.7 ) Pub Date : 2019-10-24 , DOI: 10.1039/c9ay01659g Panxue Wang 1, 2, 3, 4, 5 , Xuejie Wang 6, 7, 8, 9 , Yan Sun 6, 7, 8, 9 , Guoli Gong 6, 7, 8, 9 , Mingtao Fan 5, 9, 10, 11 , Lili He 1, 2, 3, 4
Analytical Methods ( IF 2.7 ) Pub Date : 2019-10-24 , DOI: 10.1039/c9ay01659g Panxue Wang 1, 2, 3, 4, 5 , Xuejie Wang 6, 7, 8, 9 , Yan Sun 6, 7, 8, 9 , Guoli Gong 6, 7, 8, 9 , Mingtao Fan 5, 9, 10, 11 , Lili He 1, 2, 3, 4
Affiliation
|
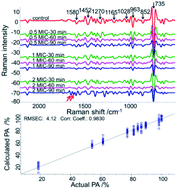
Overuse and misuse of antibiotics have resulted in wide antibiotic resistance in bacteria. In this study, surface-enhanced Raman spectroscopy (SERS) was used to rapidly characterize the responses of lactic acid bacteria (LAB) to antibiotics targeting the bacterial cell wall. Lactobacillus delbrueckii subsp. bulgaricus (Lb. bulgaricus) ATCC 11842 was used to represent LAB strains that are widely used in the food industry. Penicillin G, ampicillin and vancomycin were used to treat Lb. bulgaricus ATCC 11842 at different toxic levels for 30, 60 and 90 min, respectively. SERS spectra of Lb. bulgaricus after each treatment were collected to characterize the responses of Lb. bulgaricus. The collected spectra were analyzed by principal component analysis (PCA) and partial least squares regression (PLSR). Results suggested that all three antibiotics induced significant peak changes in LAB, and the spectral changes induced by each antibiotic treatment were significantly different. Moreover, the antibiotic-induced spectral changes in Lb. bulgaricus had good correlation with its proliferation ability and could be potentially used as a base for rapid quantification of the antibiotic susceptibility of bacteria. The developed method could detect the reaction of LAB to antibiotic treatments within 3 h. Therefore, the developed SERS method has the potential to be used to rapidly discriminate and quantify the antibiotic susceptibility of LAB. Moreover, such a rapid and direct method could contribute to microbial antibiotic susceptibility detection research.
中文翻译:

使用表面增强拉曼光谱法快速鉴定和定量乳酸菌的抗生素敏感性†
过度使用和滥用抗生素已导致细菌产生广泛的抗生素耐药性。在这项研究中,表面增强拉曼光谱(SERS)用于快速表征乳酸菌(LAB)对靶向细菌细胞壁的抗生素的反应。德氏乳杆菌亚种。保加利亚(Lb. bulgaricus)ATCC 11842用于代表在食品工业中广泛使用的LAB菌株。青霉素G,氨苄西林和万古霉素用于治疗Lb。保加利亚ATCC 11842分别在30、60和90分钟内具有不同的毒性水平。Lb的SERS光谱。每次处理后收集保加利亚以表征Lb的反应。保加利亚。收集的光谱通过主成分分析(PCA)和偏最小二乘回归(PLSR)进行分析。结果表明,所有三种抗生素均会在LAB中引起明显的峰变化,并且每种抗生素处理引起的光谱变化均存在显着差异。此外,抗生素诱导的Lb光谱变化。保加利亚与它的增殖能力具有良好的相关性,并有可能被用作快速定量测定细菌对抗生素敏感性的基础。所开发的方法可以在3小时内检测LAB对抗生素治疗的反应。因此,已开发的SERS方法具有用于快速区分和量化LAB的抗生素敏感性的潜力。而且,这种快速直接的方法可以为微生物抗生素敏感性检测研究做出贡献。
更新日期:2019-12-23
中文翻译:

使用表面增强拉曼光谱法快速鉴定和定量乳酸菌的抗生素敏感性†
过度使用和滥用抗生素已导致细菌产生广泛的抗生素耐药性。在这项研究中,表面增强拉曼光谱(SERS)用于快速表征乳酸菌(LAB)对靶向细菌细胞壁的抗生素的反应。德氏乳杆菌亚种。保加利亚(Lb. bulgaricus)ATCC 11842用于代表在食品工业中广泛使用的LAB菌株。青霉素G,氨苄西林和万古霉素用于治疗Lb。保加利亚ATCC 11842分别在30、60和90分钟内具有不同的毒性水平。Lb的SERS光谱。每次处理后收集保加利亚以表征Lb的反应。保加利亚。收集的光谱通过主成分分析(PCA)和偏最小二乘回归(PLSR)进行分析。结果表明,所有三种抗生素均会在LAB中引起明显的峰变化,并且每种抗生素处理引起的光谱变化均存在显着差异。此外,抗生素诱导的Lb光谱变化。保加利亚与它的增殖能力具有良好的相关性,并有可能被用作快速定量测定细菌对抗生素敏感性的基础。所开发的方法可以在3小时内检测LAB对抗生素治疗的反应。因此,已开发的SERS方法具有用于快速区分和量化LAB的抗生素敏感性的潜力。而且,这种快速直接的方法可以为微生物抗生素敏感性检测研究做出贡献。











































 京公网安备 11010802027423号
京公网安备 11010802027423号