当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
An omnidirectional visualization model of personalized gene regulatory networks.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-10-11 , DOI: 10.1038/s41540-019-0116-1 Chixiang Chen 1, 2 , Libo Jiang 3 , Guifang Fu 4 , Ming Wang 1, 2 , Yaqun Wang 5 , Biyi Shen 2 , Zhenqiu Liu 2 , Zuoheng Wang 6 , Wei Hou 7 , Scott A Berceli 8, 9, 10 , Rongling Wu 1, 2
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-10-11 , DOI: 10.1038/s41540-019-0116-1 Chixiang Chen 1, 2 , Libo Jiang 3 , Guifang Fu 4 , Ming Wang 1, 2 , Yaqun Wang 5 , Biyi Shen 2 , Zhenqiu Liu 2 , Zuoheng Wang 6 , Wei Hou 7 , Scott A Berceli 8, 9, 10 , Rongling Wu 1, 2
Affiliation
|
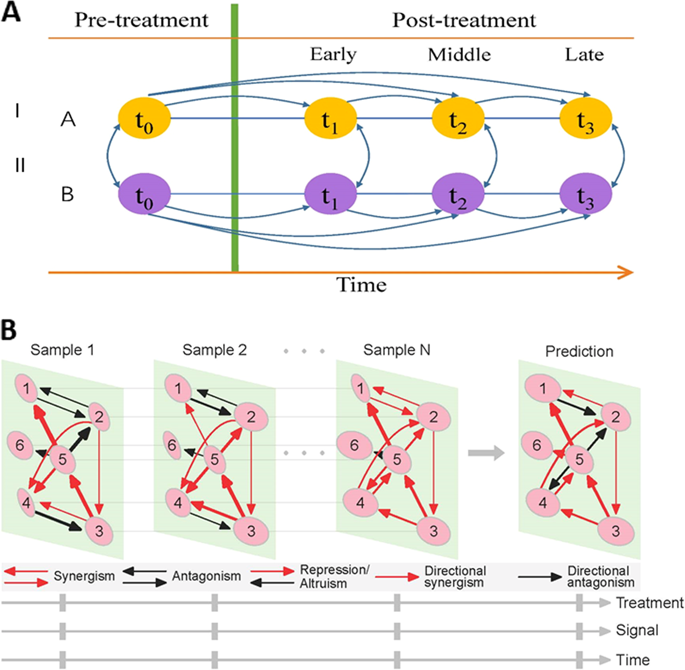
Gene regulatory networks (GRNs) have been widely used as a fundamental tool to reveal the genomic mechanisms that underlie the individual's response to environmental and developmental cues. Standard approaches infer GRNs as holistic graphs of gene co-expression, but such graphs cannot quantify how gene-gene interactions vary among individuals and how they alter structurally across spatiotemporal gradients. Here, we develop a general framework for inferring informative, dynamic, omnidirectional, and personalized networks (idopNetworks) from routine transcriptional experiments. This framework is constructed by a system of quasi-dynamic ordinary differential equations (qdODEs) derived from the combination of ecological and evolutionary theories. We reconstruct idopNetworks using genomic data from a surgical experiment and illustrate how network structure is associated with surgical response to infrainguinal vein bypass grafting and the outcome of grafting. idopNetworks may shed light on genotype-phenotype relationships and provide valuable information for personalized medicine.
中文翻译:

个性化基因调控网络的全方位可视化模型。
基因调控网络(GRN)已被广泛用作揭示个体对环境和发育线索反应的基因组机制的基本工具。标准方法将 GRN 推断为基因共表达的整体图,但此类图无法量化个体之间基因-基因相互作用如何变化以及它们如何跨时空梯度进行结构改变。在这里,我们开发了一个通用框架,用于从常规转录实验中推断信息丰富的、动态的、全向的和个性化的网络(idopNetworks)。该框架由生态和进化理论相结合的准动态常微分方程(qdODE)系统构建。我们使用手术实验的基因组数据重建了idopNetworks,并说明了网络结构如何与腹股沟下静脉旁路移植术的手术反应和移植结果相关。 idopNetworks 可以揭示基因型-表型关系,并为个性化医疗提供有价值的信息。
更新日期:2019-10-11
中文翻译:

个性化基因调控网络的全方位可视化模型。
基因调控网络(GRN)已被广泛用作揭示个体对环境和发育线索反应的基因组机制的基本工具。标准方法将 GRN 推断为基因共表达的整体图,但此类图无法量化个体之间基因-基因相互作用如何变化以及它们如何跨时空梯度进行结构改变。在这里,我们开发了一个通用框架,用于从常规转录实验中推断信息丰富的、动态的、全向的和个性化的网络(idopNetworks)。该框架由生态和进化理论相结合的准动态常微分方程(qdODE)系统构建。我们使用手术实验的基因组数据重建了idopNetworks,并说明了网络结构如何与腹股沟下静脉旁路移植术的手术反应和移植结果相关。 idopNetworks 可以揭示基因型-表型关系,并为个性化医疗提供有价值的信息。











































 京公网安备 11010802027423号
京公网安备 11010802027423号