Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Stereochemical Assessment of (φ,ψ) Outliers in Protein Structures Using Bond Geometry-Specific Ramachandran Steric-Maps.
Structure ( IF 4.4 ) Pub Date : 2019-10-10 , DOI: 10.1016/j.str.2019.09.009 Ashraya Ravikumar 1 , Chandrasekharan Ramakrishnan 1 , Narayanaswamy Srinivasan 1
Structure ( IF 4.4 ) Pub Date : 2019-10-10 , DOI: 10.1016/j.str.2019.09.009 Ashraya Ravikumar 1 , Chandrasekharan Ramakrishnan 1 , Narayanaswamy Srinivasan 1
Affiliation
|
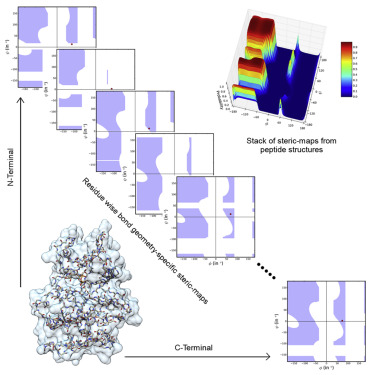
Ramachandran validation of protein structures is commonly performed using developments, such as MolProbity. We suggest tailoring such analyses by position-wise, geometry-specific steric-maps, which show (φ,ψ) regions with steric-clash at every residue position. These maps are different from the classical steric-map because they are highly sensitive to bond length and angle values that are used, in our steric-maps, as observed in the residue positions in super-high-resolution peptide and protein structures. (φ,ψ) outliers observed in such structures seldom have steric-clash. Therefore, we propose that a (φ,ψ) outlier is unacceptable if it is located within the steric-clash region of a bond geometry-specific steric-map for a residue position. These steric-maps also suggest position-specific accessible (φ,ψ) space. The PARAMA web resource performs in-depth position-wise analysis of protein structures using bond geometry-specific steric-maps.
中文翻译:

使用键几何特定的Ramachandran Steric-Maps对蛋白质结构中(φ,ψ)离群值进行立体化学评估。
Ramachandran对蛋白质结构的验证通常使用诸如MolProbity这样的开发工具来进行。我们建议通过按位置排列的,特定于几何图形的空间图来裁剪此类分析,该图显示(φ,ψ)在每个残基位置处都有空间冲突的区域。这些图谱不同于经典的空间图谱,因为它们对在我们的空间图谱中使用的键长和角度值高度敏感,正如在超高分辨率肽和蛋白质结构中的残基位置所观察到的那样。在此类结构中观察到的(φ,ψ)异常值很少具有空间冲突。因此,我们提出(φ,ψ)离群值是不可接受的,如果它位于键几何特定空间图的残基位置的空间冲突区域内。这些空间图还建议特定于位置的可访问(φ,ψ)空间。
更新日期:2019-10-10
中文翻译:

使用键几何特定的Ramachandran Steric-Maps对蛋白质结构中(φ,ψ)离群值进行立体化学评估。
Ramachandran对蛋白质结构的验证通常使用诸如MolProbity这样的开发工具来进行。我们建议通过按位置排列的,特定于几何图形的空间图来裁剪此类分析,该图显示(φ,ψ)在每个残基位置处都有空间冲突的区域。这些图谱不同于经典的空间图谱,因为它们对在我们的空间图谱中使用的键长和角度值高度敏感,正如在超高分辨率肽和蛋白质结构中的残基位置所观察到的那样。在此类结构中观察到的(φ,ψ)异常值很少具有空间冲突。因此,我们提出(φ,ψ)离群值是不可接受的,如果它位于键几何特定空间图的残基位置的空间冲突区域内。这些空间图还建议特定于位置的可访问(φ,ψ)空间。









































 京公网安备 11010802027423号
京公网安备 11010802027423号