当前位置:
X-MOL 学术
›
Acc. Chem. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Chemical Biology Approaches to Interrogate the Selenoproteome.
Accounts of Chemical Research ( IF 18.3 ) Pub Date : 2019-09-16 , DOI: 10.1021/acs.accounts.9b00379 Jennifer C Peeler 1 , Eranthie Weerapana 1
Accounts of Chemical Research ( IF 18.3 ) Pub Date : 2019-09-16 , DOI: 10.1021/acs.accounts.9b00379 Jennifer C Peeler 1 , Eranthie Weerapana 1
Affiliation
|
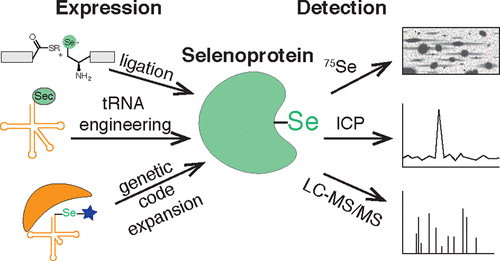
Selenoproteins are the family of proteins that contain the amino acid selenocysteine. Many selenoproteins, including glutathione peroxidases and thioredoxin reductases, play a role in maintaining cellular redox homeostasis. There are a number of examples of homologues of selenoproteins that utilize cysteine residues, raising the question of why selenocysteines are utilized. One hypothesis is that incorporation of selenocysteine protects against irreversible overoxidation, typical of cysteine-containing homologues under high oxidative stress. Studies of selenocysteine function are hampered by challenges both in detection and in recombinant expression of selenoproteins. In fact, about half of the 25 known human selenoproteins remain uncharacterized. Historically, selenoproteins were first detected via labeling with radioactive 75Se or by use of inductively coupled plasma-mass spectrometry to monitor nonradioactive selenium. More recently, tandem mass-spectrometry techniques have been developed to detect selenocysteine-containing peptides. For example, the isotopic distribution of selenium has been used as a unique signature to identify selenium-containing peptides from unenriched proteome samples. Additionally, selenocysteine-containing proteins and peptides were selectively enriched using thiol-reactive electrophiles by exploiting the increased reactivity of selenols relative to thiols, especially under low pH conditions. Importantly, the reactivity-based enrichment of selenoproteins can differentiate between oxidized and reduced selenoproteins, providing insight into the activity state. These mass spectrometry-based selenoprotein detection approaches have enabled (1) production of selenoproteome expression atlases, (2) identification of aging-associated changes in selenoprotein expression, (3) characterization of selenocysteine reactivity across the selenoprotein family, and (4) interrogation of selenoprotein targets of small-molecule drugs. Further investigations of selenoprotein function would benefit from recombinant expression of selenoproteins. However, the endogenous mechanism of selenoprotein production makes recombinant expression challenging. Primarily, selenocysteine is biosynthesized on its own tRNA, is dependent on multiple enzymatic steps, and is highly sensitive to selenium concentrations. Furthermore, selenocysteine is encoded by the stop codon UGA, and suppression of that stop codon requires a selenocysteine insertion sequence element in the selenoprotein mRNA. In order to circumvent the low efficiency of the endogenous machinery, selenoproteins have been produced in vitro through native chemical ligation and expressed protein ligation. Attempts have also been made to engineer the endogenous machinery for increased efficiency, including recoding the selenocysteine codon, and engineering the tRNA and the selenocysteine insertion sequence element. Alternatively, genetic code expansion can be used to generate selenoproteins. This approach allows for selenoprotein production directly within its native cellular environment, while bypassing the endogenous selenocysteine incorporation machinery. Furthermore, by incorporating a caged selenocysteine by genetic code expansion, selenoprotein activity can be spatially and temporally controlled. Genetic code expansion has allowed for the expression and uncaging of human selenoproteins in E. coli and more recently in mammalian cells. Together, advances in selenoprotein detection and expression should enable a better understanding of selenoprotein function and provide insight into the necessity for selenocysteine production.
中文翻译:

询问硒蛋白组的化学生物学方法。
硒蛋白是包含氨基酸硒代半胱氨酸的蛋白质家族。许多硒蛋白,包括谷胱甘肽过氧化物酶和硫氧还蛋白还原酶,都在维持细胞氧化还原稳态中发挥作用。利用半胱氨酸残基的硒蛋白同源物有许多例子,提出了为什么要使用硒代半胱氨酸的问题。一种假设是硒代半胱氨酸的掺入可防止不可逆的过氧化,这是在高氧化应激下典型的含半胱氨酸的同系物。硒代半胱氨酸功能的研究受到硒蛋白的检测和重组表达的挑战的阻碍。实际上,在25种已知的人类硒蛋白中,约有一半仍未鉴定。历史上,首先通过放射性75 Se标记或通过电感耦合等离子体质谱法监测非放射性硒来检测硒蛋白。最近,已经开发了串联质谱技术来检测含硒代半胱氨酸的肽。例如,硒的同位素分布已被用作从未富集的蛋白质组样品中鉴定含硒肽的独特标记。另外,通过利用硒醇相对于硫醇的增加的反应性,特别是在低pH条件下,使用硫醇反应性的亲电试剂选择性地富集含硒代半胱氨酸的蛋白质和肽。重要的是,基于反应性的硒蛋白富集可以区分氧化硒蛋白和还原硒蛋白,从而洞悉活性状态。这些基于质谱的硒蛋白检测方法能够(1)生成硒蛋白表达图谱,(2)识别与老化相关的硒蛋白表达变化,(3)表征整个硒蛋白家族中的硒代半胱氨酸反应性,和(4)讯问小分子药物的硒蛋白靶标。硒蛋白功能的进一步研究将受益于硒蛋白的重组表达。然而,硒蛋白产生的内源性机制使重组表达具有挑战性。首先,硒代半胱氨酸是通过自身的tRNA进行生物合成的,它依赖于多个酶促步骤,并且对硒浓度高度敏感。此外,硒代半胱氨酸由终止密码子UGA编码,并且抑制该终止密码子需要硒蛋白mRNA中的硒代半胱氨酸插入序列元件。为了避免内源性机器的低效率,已经通过天然化学连接和表达的蛋白质连接在体外产生了硒蛋白。还尝试了改造内源机制以提高效率,包括重新编码硒代半胱氨酸密码子,以及改造tRNA和硒代半胱氨酸插入序列元件。或者,遗传密码扩展可用于产生硒蛋白。这种方法可以直接在其天然细胞环境中产生硒蛋白,同时绕过内源性硒代半胱氨酸掺入机制。此外,通过通过遗传密码扩展并入笼状硒代半胱氨酸,硒蛋白活性可以在空间和时间上得到控制。遗传密码的扩展允许人类硒蛋白在大肠杆菌中和最近在哺乳动物细胞中的表达和释放。在一起,硒蛋白检测和表达的进展应能使人们更好地了解硒蛋白的功能,并深入了解硒代半胱氨酸生产的必要性。
更新日期:2019-09-16
中文翻译:

询问硒蛋白组的化学生物学方法。
硒蛋白是包含氨基酸硒代半胱氨酸的蛋白质家族。许多硒蛋白,包括谷胱甘肽过氧化物酶和硫氧还蛋白还原酶,都在维持细胞氧化还原稳态中发挥作用。利用半胱氨酸残基的硒蛋白同源物有许多例子,提出了为什么要使用硒代半胱氨酸的问题。一种假设是硒代半胱氨酸的掺入可防止不可逆的过氧化,这是在高氧化应激下典型的含半胱氨酸的同系物。硒代半胱氨酸功能的研究受到硒蛋白的检测和重组表达的挑战的阻碍。实际上,在25种已知的人类硒蛋白中,约有一半仍未鉴定。历史上,首先通过放射性75 Se标记或通过电感耦合等离子体质谱法监测非放射性硒来检测硒蛋白。最近,已经开发了串联质谱技术来检测含硒代半胱氨酸的肽。例如,硒的同位素分布已被用作从未富集的蛋白质组样品中鉴定含硒肽的独特标记。另外,通过利用硒醇相对于硫醇的增加的反应性,特别是在低pH条件下,使用硫醇反应性的亲电试剂选择性地富集含硒代半胱氨酸的蛋白质和肽。重要的是,基于反应性的硒蛋白富集可以区分氧化硒蛋白和还原硒蛋白,从而洞悉活性状态。这些基于质谱的硒蛋白检测方法能够(1)生成硒蛋白表达图谱,(2)识别与老化相关的硒蛋白表达变化,(3)表征整个硒蛋白家族中的硒代半胱氨酸反应性,和(4)讯问小分子药物的硒蛋白靶标。硒蛋白功能的进一步研究将受益于硒蛋白的重组表达。然而,硒蛋白产生的内源性机制使重组表达具有挑战性。首先,硒代半胱氨酸是通过自身的tRNA进行生物合成的,它依赖于多个酶促步骤,并且对硒浓度高度敏感。此外,硒代半胱氨酸由终止密码子UGA编码,并且抑制该终止密码子需要硒蛋白mRNA中的硒代半胱氨酸插入序列元件。为了避免内源性机器的低效率,已经通过天然化学连接和表达的蛋白质连接在体外产生了硒蛋白。还尝试了改造内源机制以提高效率,包括重新编码硒代半胱氨酸密码子,以及改造tRNA和硒代半胱氨酸插入序列元件。或者,遗传密码扩展可用于产生硒蛋白。这种方法可以直接在其天然细胞环境中产生硒蛋白,同时绕过内源性硒代半胱氨酸掺入机制。此外,通过通过遗传密码扩展并入笼状硒代半胱氨酸,硒蛋白活性可以在空间和时间上得到控制。遗传密码的扩展允许人类硒蛋白在大肠杆菌中和最近在哺乳动物细胞中的表达和释放。在一起,硒蛋白检测和表达的进展应能使人们更好地了解硒蛋白的功能,并深入了解硒代半胱氨酸生产的必要性。

































 京公网安备 11010802027423号
京公网安备 11010802027423号