当前位置:
X-MOL 学术
›
Mol. Omics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Surpassing 10 000 identified and quantified proteins in a single run by optimizing current LC-MS instrumentation and data analysis strategy.
Molecular Omics ( IF 3.0 ) Pub Date : 2019-10-07 , DOI: 10.1039/c9mo00082h Jan Muntel 1 , Tejas Gandhi , Lynn Verbeke , Oliver M Bernhardt , Tobias Treiber , Roland Bruderer , Lukas Reiter
Molecular Omics ( IF 3.0 ) Pub Date : 2019-10-07 , DOI: 10.1039/c9mo00082h Jan Muntel 1 , Tejas Gandhi , Lynn Verbeke , Oliver M Bernhardt , Tobias Treiber , Roland Bruderer , Lukas Reiter
Affiliation
|
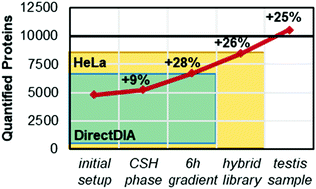
Comprehensive proteome quantification is crucial for a better understanding of underlying mechanisms of diseases. Liquid chromatography mass spectrometry (LC-MS) has become the method of choice for comprehensive proteome quantification due to its power and versatility. Even though great advances have been made in recent years, full proteome coverage for complex samples remains challenging due to the high dynamic range of protein expression. Additionally, when studying disease regulatory proteins, biomarkers or potential drug targets are often low abundant, such as for instance kinases and transcription factors. Here, we show that with improvements in chromatography and data analysis the single shot proteome coverage can go beyond 10 000 proteins in human tissue. In a testis cancer study, we quantified 11 200 proteins using data independent acquisition (DIA). This depth was achieved with a false discovery rate of 1% which was experimentally validated using a two species test. We introduce the concept of hybrid libraries which combines the strength of direct searching of DIA data as well as the use of large project-specific or published DDA data sets. Remarkably deep proteome coverage is possible using hybrid libraries without the additional burden of creating a project-specific library. Within the testis cancer set, we found a large proportion of proteins in an altered expression (in total: 3351; 1453 increased in cancer). Many of these proteins could be linked to the hallmarks of cancer. For example, the complement system was downregulated which helps to evade the immune response and chromosomal replication was upregulated indicating a dysregulated cell cycle.
中文翻译:

通过优化当前的LC-MS仪器和数据分析策略,单次运行即可识别超过10,000种蛋白质。
全面的蛋白质组定量对于更好地了解疾病的潜在机制至关重要。液相色谱质谱法(LC-MS)由于其强大的功能和多功能性,已成为蛋白质组学定量分析的首选方法。尽管近年来取得了长足的进步,但是由于蛋白质表达的高动态范围,复杂样品的完整蛋白质组覆盖仍然具有挑战性。另外,当研究疾病调节蛋白时,生物标志物或潜在的药物靶标常常是低丰度的,例如激酶和转录因子。在这里,我们表明,随着色谱和数据分析的改进,单发蛋白质组的覆盖范围可以超过人体组织中的10 000种蛋白质。在睾丸癌研究中,我们使用数据独立采集(DIA)量化了11200种蛋白质。该深度是通过1%的错误发现率实现的,该错误发现率已通过两次物种测试进行了实验验证。我们介绍了混合库的概念,它结合了直接搜索DIA数据以及使用大型项目特定或已发布DDA数据集的优势。使用混合库可以实现非常深的蛋白质组覆盖,而不会增加创建特定于项目的库的负担。在睾丸癌中,我们发现表达改变的蛋白质占很大比例(总计:3351;癌症中增加了1453)。这些蛋白质中的许多可能与癌症的特征有关。例如,
更新日期:2019-10-08
中文翻译:

通过优化当前的LC-MS仪器和数据分析策略,单次运行即可识别超过10,000种蛋白质。
全面的蛋白质组定量对于更好地了解疾病的潜在机制至关重要。液相色谱质谱法(LC-MS)由于其强大的功能和多功能性,已成为蛋白质组学定量分析的首选方法。尽管近年来取得了长足的进步,但是由于蛋白质表达的高动态范围,复杂样品的完整蛋白质组覆盖仍然具有挑战性。另外,当研究疾病调节蛋白时,生物标志物或潜在的药物靶标常常是低丰度的,例如激酶和转录因子。在这里,我们表明,随着色谱和数据分析的改进,单发蛋白质组的覆盖范围可以超过人体组织中的10 000种蛋白质。在睾丸癌研究中,我们使用数据独立采集(DIA)量化了11200种蛋白质。该深度是通过1%的错误发现率实现的,该错误发现率已通过两次物种测试进行了实验验证。我们介绍了混合库的概念,它结合了直接搜索DIA数据以及使用大型项目特定或已发布DDA数据集的优势。使用混合库可以实现非常深的蛋白质组覆盖,而不会增加创建特定于项目的库的负担。在睾丸癌中,我们发现表达改变的蛋白质占很大比例(总计:3351;癌症中增加了1453)。这些蛋白质中的许多可能与癌症的特征有关。例如,









































 京公网安备 11010802027423号
京公网安备 11010802027423号