当前位置:
X-MOL 学术
›
Toxicol. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Methods for the analysis of transcriptome dynamics.
Toxicology Research ( IF 2.2 ) Pub Date : 2019-07-26 00:00:00 , DOI: 10.1039/c9tx00088g Daniela F Rodrigues 1 , Vera M Costa 1 , Ricardo Silvestre 2, 3 , Maria L Bastos 1 , Félix Carvalho 1
Toxicology Research ( IF 2.2 ) Pub Date : 2019-07-26 00:00:00 , DOI: 10.1039/c9tx00088g Daniela F Rodrigues 1 , Vera M Costa 1 , Ricardo Silvestre 2, 3 , Maria L Bastos 1 , Félix Carvalho 1
Affiliation
|
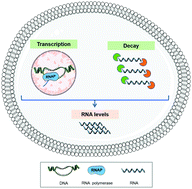
The transcriptome is the complete set of transcripts in a cell or tissue and includes ribosomal RNA (rRNA), messenger RNA (mRNA), transfer RNA (tRNA), and regulatory noncoding RNA. At steady-state, the transcriptome results from a compensatory variation of the transcription and decay rate to maintain the RNA concentration constant. RNA transcription constitutes the first stage in gene expression, and thus is a major and primary mode of gene expression control. Nevertheless, regulation of RNA decay is also a key factor in gene expression control, involving either selective RNA stabilization or enhanced degradation. Transcriptome analysis allows the identification of gene expression alterations, providing new insights regarding the pathways and mechanisms involved in physiological and pathological processes. Upon perturbation of cell homeostasis, rapid changes in gene expression are required to adapt to new conditions. Thus, to better understand the regulatory mechanisms associated with gene expression alterations, it is vital to acknowledge the relative contribution of RNA synthesis and decay to the transcriptome. To the toxicology field, the study of gene expression regulation mechanisms can help identify the early and mechanistic relevant cellular events associated with a particular response. This review aims to provide a critical comparison of the available methods used to analyze the contribution of RNA transcription and decay to gene expression dynamics. Notwithstanding, an integration of the data obtained is necessary to understand the entire repercussions of gene transcription changes at a system-level. Thus, a brief overview of the methods available for the integration and analysis of the data obtained from transcriptome analysis will also be provided.
中文翻译:

转录组动力学分析方法。
转录组是细胞或组织中完整的转录组,包括核糖体RNA(rRNA),信使RNA(mRNA),转移RNA(tRNA)和调节性非编码RNA。在稳态下,转录组是由转录和衰变速率的补偿性变化产生的,以保持RNA浓度恒定。RNA转录构成基因表达的第一阶段,因此是基因表达控制的主要方式。尽管如此,RNA衰变的调控也是基因表达控制的关键因素,涉及选择性的RNA稳定或增强的降解。转录组分析可以鉴定基因表达的变化,从而提供有关生理和病理过程涉及的途径和机制的新见解。扰动细胞稳态后,需要基因表达的快速变化以适应新的条件。因此,为了更好地理解与基因表达改变相关的调控机制,至关重要的是要认识到RNA合成和衰变对转录组的相对贡献。对于毒理学领域,基因表达调控机制的研究可以帮助识别与特定反应相关的早期和机制相关的细胞事件。这篇综述旨在提供可用于分析RNA转录和衰变对基因表达动力学的影响的可用方法的重要比较。尽管如此,获得的数据的整合对于理解系统级基因转录变化的整个影响是必要的。因此,
更新日期:2019-07-26
中文翻译:

转录组动力学分析方法。
转录组是细胞或组织中完整的转录组,包括核糖体RNA(rRNA),信使RNA(mRNA),转移RNA(tRNA)和调节性非编码RNA。在稳态下,转录组是由转录和衰变速率的补偿性变化产生的,以保持RNA浓度恒定。RNA转录构成基因表达的第一阶段,因此是基因表达控制的主要方式。尽管如此,RNA衰变的调控也是基因表达控制的关键因素,涉及选择性的RNA稳定或增强的降解。转录组分析可以鉴定基因表达的变化,从而提供有关生理和病理过程涉及的途径和机制的新见解。扰动细胞稳态后,需要基因表达的快速变化以适应新的条件。因此,为了更好地理解与基因表达改变相关的调控机制,至关重要的是要认识到RNA合成和衰变对转录组的相对贡献。对于毒理学领域,基因表达调控机制的研究可以帮助识别与特定反应相关的早期和机制相关的细胞事件。这篇综述旨在提供可用于分析RNA转录和衰变对基因表达动力学的影响的可用方法的重要比较。尽管如此,获得的数据的整合对于理解系统级基因转录变化的整个影响是必要的。因此,









































 京公网安备 11010802027423号
京公网安备 11010802027423号