当前位置:
X-MOL 学术
›
Nat. Protoc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis.
Nature Protocols ( IF 14.8 ) Pub Date : 2019-07-24 , DOI: 10.1038/s41596-019-0195-x Shuqiang Li 1, 2, 3 , Jing Sun 2 , Rosa Allesøe 2, 4 , Krishnalekha Datta 5 , Yun Bao 5 , Giacomo Oliveira 2 , Juliet Forman 1, 2, 3 , Roger Jin 6 , Lars Rønn Olsen 4 , Derin B Keskin 2, 3 , Sachet A Shukla 2, 3 , Catherine J Wu 1, 2 , Kenneth J Livak 2, 3
Nature Protocols ( IF 14.8 ) Pub Date : 2019-07-24 , DOI: 10.1038/s41596-019-0195-x Shuqiang Li 1, 2, 3 , Jing Sun 2 , Rosa Allesøe 2, 4 , Krishnalekha Datta 5 , Yun Bao 5 , Giacomo Oliveira 2 , Juliet Forman 1, 2, 3 , Roger Jin 6 , Lars Rønn Olsen 4 , Derin B Keskin 2, 3 , Sachet A Shukla 2, 3 , Catherine J Wu 1, 2 , Kenneth J Livak 2, 3
Affiliation
|
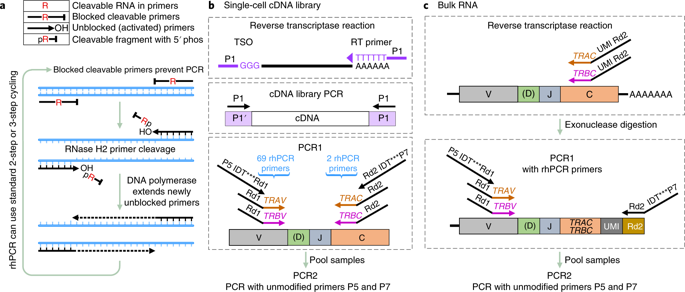
RNase H-dependent PCR-enabled T-cell receptor sequencing (rhTCRseq) can be used to determine paired alpha/beta T-cell receptor (TCR) clonotypes in single cells or perform alpha and beta TCR repertoire analysis in bulk RNA samples. With the enhanced specificity of RNase H-dependent PCR (rhPCR), it achieves TCR-specific amplification and addition of dual-index barcodes in a single PCR step. For single cells, the protocol includes sorting of single cells into plates, generation of cDNA libraries, a TCR-specific amplification step, a second PCR on pooled sample to generate a sequencing library, and sequencing. In the bulk method, sorting and cDNA library steps are replaced with a reverse-transcriptase (RT) reaction that adds a unique molecular identifier (UMI) to each cDNA molecule to improve the accuracy of repertoire-frequency measurements. Compared to other methods for TCR sequencing, rhTCRseq has a streamlined workflow and the ability to analyze single cells in 384-well plates. Compared to TCR reconstruction from single-cell transcriptome sequencing data, it improves the success rate for obtaining paired alpha/beta information and ensures recovery of complete complementarity-determining region 3 (CDR3) sequences, a prerequisite for cloning/expression of discovered TCRs. Although it has lower throughput than droplet-based methods, rhTCRseq is well-suited to analysis of small sorted populations, especially when analysis of 96 or 384 single cells is sufficient to identify predominant T-cell clones. For single cells, sorting typically requires 2-4 h and can be performed days, or even months, before library construction and data processing, which takes ~4 d; the bulk RNA protocol takes ~3 d.
中文翻译:

RNase H 依赖性 PCR 启用的 T 细胞受体测序,可对 T 细胞受体 mRNA 进行高度特异性和高效的靶向测序,用于单细胞和库分析。
RNase H 依赖性 PCR 启用的 T 细胞受体测序 (rhTCRseq) 可用于确定单个细胞中的配对 α/β T 细胞受体 (TCR) 克隆型,或对大量 RNA 样本进行 α 和β TCR 谱分析。借助RNase H-dependent PCR (rhPCR)增强的特异性,它在单个PCR步骤中实现了TCR特异性扩增和双索引条码的添加。对于单个细胞,该协议包括将单个细胞分选到板中、生成 cDNA 文库、TCR 特定扩增步骤、对合并样本进行第二次 PCR 以生成测序文库和测序。在批量方法中,分选和 cDNA 文库步骤被逆转录酶 (RT) 反应所取代,该反应为每个 cDNA 分子添加一个独特的分子标识符 (UMI),以提高库频率测量的准确性。与其他 TCR 测序方法相比,rhTCRseq 具有简化的工作流程和分析 384 孔板中单个细胞的能力。与从单细胞转录组测序数据重建 TCR 相比,它提高了获得配对 alpha/beta 信息的成功率,并确保恢复完整的互补决定区 3 (CDR3) 序列,这是已发现的 TCR 克隆/表达的先决条件。尽管它的通量低于基于液滴的方法,但 rhTCRseq 非常适合分析小分类群体,尤其是当分析 96 或 384 个单细胞足以识别主要 T 细胞克隆时。对于单个细胞,排序通常需要 2-4 小时,并且可以在建库和数据处理之前进行数天甚至数月,大约需要 4 天;
更新日期:2019-11-18
中文翻译:

RNase H 依赖性 PCR 启用的 T 细胞受体测序,可对 T 细胞受体 mRNA 进行高度特异性和高效的靶向测序,用于单细胞和库分析。
RNase H 依赖性 PCR 启用的 T 细胞受体测序 (rhTCRseq) 可用于确定单个细胞中的配对 α/β T 细胞受体 (TCR) 克隆型,或对大量 RNA 样本进行 α 和β TCR 谱分析。借助RNase H-dependent PCR (rhPCR)增强的特异性,它在单个PCR步骤中实现了TCR特异性扩增和双索引条码的添加。对于单个细胞,该协议包括将单个细胞分选到板中、生成 cDNA 文库、TCR 特定扩增步骤、对合并样本进行第二次 PCR 以生成测序文库和测序。在批量方法中,分选和 cDNA 文库步骤被逆转录酶 (RT) 反应所取代,该反应为每个 cDNA 分子添加一个独特的分子标识符 (UMI),以提高库频率测量的准确性。与其他 TCR 测序方法相比,rhTCRseq 具有简化的工作流程和分析 384 孔板中单个细胞的能力。与从单细胞转录组测序数据重建 TCR 相比,它提高了获得配对 alpha/beta 信息的成功率,并确保恢复完整的互补决定区 3 (CDR3) 序列,这是已发现的 TCR 克隆/表达的先决条件。尽管它的通量低于基于液滴的方法,但 rhTCRseq 非常适合分析小分类群体,尤其是当分析 96 或 384 个单细胞足以识别主要 T 细胞克隆时。对于单个细胞,排序通常需要 2-4 小时,并且可以在建库和数据处理之前进行数天甚至数月,大约需要 4 天;



























 京公网安备 11010802027423号
京公网安备 11010802027423号