当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
CHARMM‐GUI DEER facilitator for spin‐pair distance distribution calculations and preparation of restrained‐ensemble molecular dynamics simulations
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2019-07-22 , DOI: 10.1002/jcc.26032 Yifei Qi 1 , Jumin Lee 2 , Xi Cheng 3 , Rong Shen 4 , Shahidul M Islam 5 , Benoît Roux 4 , Wonpil Im 2
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2019-07-22 , DOI: 10.1002/jcc.26032 Yifei Qi 1 , Jumin Lee 2 , Xi Cheng 3 , Rong Shen 4 , Shahidul M Islam 5 , Benoît Roux 4 , Wonpil Im 2
Affiliation
|
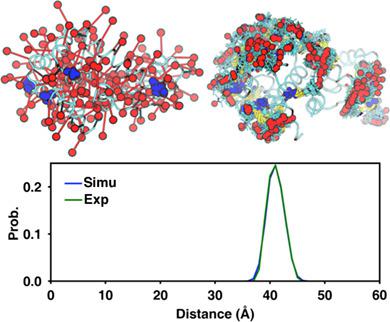
The double electron–electron resonance (DEER) is a powerful structural biology technique to obtain distance information in the range of 18 to 80 å by measuring the dipolar coupling between two unpaired electron spins. The distance distributions obtained from the experiment provide valuable structural information about the protein in its native environment that can be exploited using restrained ensemble molecular dynamics (reMD) simulations. We present a new tool DEER Facilitator in CHARMM‐GUI that consists of two modules Spin‐Pair Distributor and reMD Prepper to setup simulations that utilize information from DEER experiments. Spin‐Pair Distributor provides a web‐based interface to calculate the spin‐pair distance distribution of labeled sites in a protein using MD simulations. The calculated distribution can be used to guide the selection of the labeling sites in experiments as well as validate different protein structure models. reMD Prepper facilities the setup of reMD simulations using different types of spin labels in four different environments including vacuum, solution, micelle, and bilayer. The applications of these two modules are demonstrated with several test cases. Spin‐Pair Distributor and reMD Prepper are available at http://www.charmm-gui.org/input/deer and http://www.charmm-gui.org/input/deerre. DEER Facilitator is expected to facilitate advanced biomolecular modeling and simulation, thereby leading to an improved understanding of the structure and dynamics of complex biomolecular systems based on experimental DEER data. © 2019 Wiley Periodicals, Inc.
中文翻译:

CHARMM-GUI DEER 促进器用于自旋对距离分布计算和约束集合分子动力学模拟的准备
双电子-电子共振 (DEER) 是一种强大的结构生物学技术,通过测量两个不成对电子自旋之间的偶极耦合来获得 18 到 80 å 范围内的距离信息。从实验中获得的距离分布提供了有关蛋白质在其天然环境中的有价值的结构信息,可以使用约束集合分子动力学 (reMD) 模拟来利用这些信息。我们在 CHARMM-GUI 中展示了一个新工具 DEER Facilitator,它由两个模块 Spin-Pair Distributor 和 reMD Prepper 组成,以设置利用来自 DEER 实验的信息的模拟。Spin-Pair Distributor 提供了一个基于 Web 的界面,用于使用 MD 模拟计算蛋白质中标记位点的自旋对距离分布。计算的分布可用于指导实验中标记位点的选择以及验证不同的蛋白质结构模型。reMD Prepper 在真空、溶液、胶束和双层等四种不同环境中使用不同类型的自旋标签设置 reMD 模拟。通过几个测试用例演示了这两个模块的应用。Spin-Pair Distributor 和 reMD Prepper 可从 http://www.charmm-gui.org/input/deer 和 http://www.charmm-gui.org/input/deerre 获得。DEER Facilitator 有望促进先进的生物分子建模和模拟,从而提高对基于实验性 DEER 数据的复杂生物分子系统的结构和动力学的理解。© 2019 威利期刊公司。
更新日期:2019-07-22
中文翻译:

CHARMM-GUI DEER 促进器用于自旋对距离分布计算和约束集合分子动力学模拟的准备
双电子-电子共振 (DEER) 是一种强大的结构生物学技术,通过测量两个不成对电子自旋之间的偶极耦合来获得 18 到 80 å 范围内的距离信息。从实验中获得的距离分布提供了有关蛋白质在其天然环境中的有价值的结构信息,可以使用约束集合分子动力学 (reMD) 模拟来利用这些信息。我们在 CHARMM-GUI 中展示了一个新工具 DEER Facilitator,它由两个模块 Spin-Pair Distributor 和 reMD Prepper 组成,以设置利用来自 DEER 实验的信息的模拟。Spin-Pair Distributor 提供了一个基于 Web 的界面,用于使用 MD 模拟计算蛋白质中标记位点的自旋对距离分布。计算的分布可用于指导实验中标记位点的选择以及验证不同的蛋白质结构模型。reMD Prepper 在真空、溶液、胶束和双层等四种不同环境中使用不同类型的自旋标签设置 reMD 模拟。通过几个测试用例演示了这两个模块的应用。Spin-Pair Distributor 和 reMD Prepper 可从 http://www.charmm-gui.org/input/deer 和 http://www.charmm-gui.org/input/deerre 获得。DEER Facilitator 有望促进先进的生物分子建模和模拟,从而提高对基于实验性 DEER 数据的复杂生物分子系统的结构和动力学的理解。© 2019 威利期刊公司。











































 京公网安备 11010802027423号
京公网安备 11010802027423号