当前位置:
X-MOL 学术
›
Sleep Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
TOX3 gene variant could be associated with painful restless legs.
Sleep Medicine ( IF 3.8 ) Pub Date : 2019-07-11 , DOI: 10.1016/j.sleep.2019.07.003 Elias G Karroum 1 , Prabhjyot S Saini 2 , Lynn M Trotti 2 , David B Rye 2
Sleep Medicine ( IF 3.8 ) Pub Date : 2019-07-11 , DOI: 10.1016/j.sleep.2019.07.003 Elias G Karroum 1 , Prabhjyot S Saini 2 , Lynn M Trotti 2 , David B Rye 2
Affiliation
|
BACKGROUND/OBJECTIVE
Restless legs syndrome (RLS) is a neurological disorder with a strong genetic susceptibility. A painful RLS sub-phenotype has been described previously but the neurobiological basis for this phenotypic variant remains unknown. This study investigated whether any of the six initially discovered genomic loci associating with RLS (BTBD9, MEIS1, PTPRD, MAP2K5/SKOR1, TOX3, and an intergenic region on chromosome 2), were more strongly associated with complaints of painful versus non-painful RLS.
METHODS
RLS patients (N = 199; Age = 53.1 ± 16.8; 100% Caucasians; 57% women) diagnosed clinically were genotyped for known variants associating with RLS. Definition of painful RLS required that subjects selected "painful" from a list of 14 adjectives to describe their RLS sensory experience and answered positively to a separate question that queried specifically as to whether they perceived their RLS sensations as painful. Genotype association tests employed logistic regression analyses with assumption of an additive genetic model. Analyses were performed using PLINK software v1.07.
RESULTS
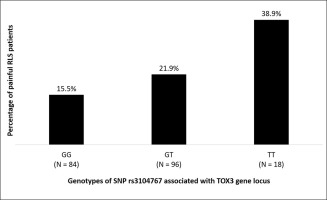
We identified two RLS patient subgroups: a painful (n = 41) and non-painful (n = 158). Among 10 tested SNPs, only rs3104767 (related to the TOX3 gene locus) was more associated with painful RLS. The minor allele T of SNP rs3104767 was associated with an increased risk of RLS being perceived as painful with an OR of 1.67 [CI = (1.01-2.74); p = 0.049]. Notably, this minor T allele associated with pain sensation in RLS patients in this study was the non-risk allele for RLS in the original RLS genome wide association study, but a similar trend was observed in a recent Parkinson disease sample study.
CONCLUSION
This study might suggest the TOX3 gene variant as a potential genetic substrate for the painful RLS sub-phenotype. This was an exploratory small study and correction for multiple comparisons would have rendered the results not significant. Therefore, the above findings require replication in larger clinical as well as population-based samples of RLS subjects.
中文翻译:

TOX3基因变异可能与腿部躁动不安有关。
背景/目的不安腿综合症(RLS)是一种遗传学易感性强的神经系统疾病。先前已经描述了痛苦的RLS亚型,但是该表型变异的神经生物学基础仍然未知。这项研究调查了最初发现的与RLS相关的六个基因组位点(BTBD9,MEIS1,PTPRD,MAP2K5 / SKOR1,TOX3和2号染色体上的一个基因间区域)中的任何一个是否与疼痛性或非疼痛性RLS的主诉相关性更强。 。方法对临床诊断为RLS的患者(N = 199;年龄= 53.1±16.8; 100%的白种人; 57%的女性)进行基因分型,以了解与RLS相关的已知变异。痛苦的RLS的定义要求受试者选择“痛苦的” 从14个形容词的列表中描述了他们的RLS感觉体验,并积极回答了一个单独的问题,该问题专门询问他们是否认为自己的RLS感觉很痛苦。基因型关联测试采用逻辑回归分析,并假设存在附加遗传模型。使用PLINK软件v1.07进行分析。结果我们确定了两个RLS患者亚组:疼痛(n = 41)和非疼痛(n = 158)。在10个经过测试的SNP中,只有rs3104767(与TOX3基因位点有关)与疼痛性RLS的关联更大。SNP rs3104767的次要等位基因T与RLS被认为是痛苦的风险增加有关,OR为1.67 [CI =(1.01-2.74);p = 0.049]。尤其,在本研究中,该与RLS患者疼痛感相关的次要T等位基因是原始RLS基因组广泛关联研究中RLS的非风险等位基因,但在最近的帕金森病样本研究中观察到了类似的趋势。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行更正会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行校正可能会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行校正可能会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。
更新日期:2019-07-11
中文翻译:

TOX3基因变异可能与腿部躁动不安有关。
背景/目的不安腿综合症(RLS)是一种遗传学易感性强的神经系统疾病。先前已经描述了痛苦的RLS亚型,但是该表型变异的神经生物学基础仍然未知。这项研究调查了最初发现的与RLS相关的六个基因组位点(BTBD9,MEIS1,PTPRD,MAP2K5 / SKOR1,TOX3和2号染色体上的一个基因间区域)中的任何一个是否与疼痛性或非疼痛性RLS的主诉相关性更强。 。方法对临床诊断为RLS的患者(N = 199;年龄= 53.1±16.8; 100%的白种人; 57%的女性)进行基因分型,以了解与RLS相关的已知变异。痛苦的RLS的定义要求受试者选择“痛苦的” 从14个形容词的列表中描述了他们的RLS感觉体验,并积极回答了一个单独的问题,该问题专门询问他们是否认为自己的RLS感觉很痛苦。基因型关联测试采用逻辑回归分析,并假设存在附加遗传模型。使用PLINK软件v1.07进行分析。结果我们确定了两个RLS患者亚组:疼痛(n = 41)和非疼痛(n = 158)。在10个经过测试的SNP中,只有rs3104767(与TOX3基因位点有关)与疼痛性RLS的关联更大。SNP rs3104767的次要等位基因T与RLS被认为是痛苦的风险增加有关,OR为1.67 [CI =(1.01-2.74);p = 0.049]。尤其,在本研究中,该与RLS患者疼痛感相关的次要T等位基因是原始RLS基因组广泛关联研究中RLS的非风险等位基因,但在最近的帕金森病样本研究中观察到了类似的趋势。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行更正会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行校正可能会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。结论这项研究可能提示TOX3基因变异可能是疼痛性RLS亚型的潜在遗传底物。这是一项探索性的小型研究,对多个比较进行校正可能会使结果不显着。因此,以上发现需要在更大的临床以及基于人群的RLS受试者样本中进行复制。









































 京公网安备 11010802027423号
京公网安备 11010802027423号