npj Biofilms and Microbiomes ( IF 7.8 ) Pub Date : 2019-06-25 , DOI: 10.1038/s41522-019-0090-9 Shi Ming Tan , Pui Yi Maria Yung , Paul E. Hutchinson , Chao Xie , Guo Hui Teo , Muhammad Hafiz Ismail , Daniela I. Drautz-Moses , Peter F. R Little , Rohan B. H. Williams , Yehuda Cohen
|
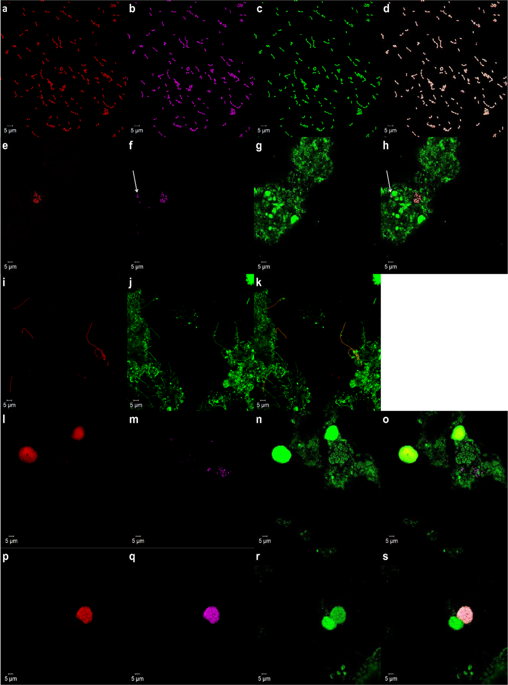
Methods for the study of member species in complex microbial communities remain a high priority, particularly for rare and/or novel member species that might play an important ecological role. Specifically, methods that link genomic information of member species with its spatial structure are lacking. This study adopts an integrative workflow that permits the characterisation of previously unclassified bacterial taxa from microbiomes through: (1) imaging of the spatial structure; (2) taxonomic classification and (3) genome recovery. Our study attempts to bridge the gaps between metagenomics/metatranscriptomics and high-resolution biomass imaging methods by developing new fluorescence in situ hybridisation (FISH) probes—termed as R-Probes—from shotgun reads that harbour hypervariable regions of the 16S rRNA gene. The sample-centric design of R-Probes means that probes can directly hybridise to OTUs as detected in shotgun sequencing surveys. The primer-free probe design captures larger microbial diversity as compared to canonical probes. R-Probes were designed from deep-sequenced RNA-Seq datasets for both FISH imaging and FISH–Fluorescence activated cell sorting (FISH–FACS). FISH–FACS was used for target enrichment of previously unclassified bacterial taxa prior to downstream multiple displacement amplification (MDA), genomic sequencing and genome recovery. After validation of the workflow on an axenic isolate of Thauera species, the techniques were applied to investigate two previously uncharacterised taxa from a tropical full-scale activated sludge community. In some instances, probe design on the hypervariable region allowed differentiation to the species level. Collectively, the workflow can be readily applied to microbiomes for which shotgun nucleic acid survey data is available.
中文翻译:

来自宏基因组学/元转录组学数据的无引物FISH探针可用于研究复杂微生物群落中未表征的分类群。
在复杂的微生物群落中研究成员物种的方法仍然是重中之重,特别是对于可能发挥重要生态作用的稀有和/或新颖成员物种而言。具体而言,缺乏将成员物种的基因组信息与其空间结构联系起来的方法。这项研究采用了一种集成的工作流程,该流程可以通过以下方式从微生物群落中表征先前未分类的细菌类群:(1)空间结构成像;(2)分类学分类和(3)基因组恢复。我们的研究尝试通过开发带有16S rRNA基因高变区的shot弹枪读数的新型荧光原位杂交(FISH)探针(称为R-Probes)来弥补宏基因组学/超基因组学与高分辨率生物质成像方法之间的差距。R-Probes的以样品为中心的设计意味着探针可以直接与shot弹枪测序调查中检测到的OTU杂交。与标准探针相比,无引物探针设计可捕获更大的微生物多样性。R-Probes是根据深度测序的RNA-Seq数据集设计的,用于FISH成像和FISH-荧光激活细胞分选(FISH-FACS)。FISH–FACS用于之前未分类细菌类群的靶标富集,然后进行下游多置换扩增(MDA),基因组测序和基因组恢复。验证工作流程后,将其无菌隔离 R-Probes是根据深度测序的RNA-Seq数据集设计的,用于FISH成像和FISH-荧光激活细胞分选(FISH-FACS)。FISH–FACS用于之前未分类细菌类群的靶标富集,然后进行下游多置换扩增(MDA),基因组测序和基因组恢复。验证工作流程后,将其无菌隔离 R-Probes是根据深度测序的RNA-Seq数据集设计的,用于FISH成像和FISH-荧光激活细胞分选(FISH-FACS)。FISH–FACS用于之前未分类细菌类群的靶标富集,然后进行下游多置换扩增(MDA),基因组测序和基因组恢复。验证工作流程后,将其无菌隔离Thauera物种,该技术被用于调查热带全尺寸活性污泥群落中两个以前未表征的分类单元。在某些情况下,高变区上的探针设计允许分化到物种水平。总体而言,该工作流程可轻松应用于散弹枪核酸调查数据可用的微生物群落。











































 京公网安备 11010802027423号
京公网安备 11010802027423号