npj Computational Materials ( IF 9.4 ) Pub Date : 2019-06-13 , DOI: 10.1038/s41524-019-0202-3 Artem A. Trofimov , Alison A. Pawlicki , Nikolay Borodinov , Shovon Mandal , Teresa J. Mathews , Mark Hildebrand , Maxim A. Ziatdinov , Katherine A. Hausladen , Paulina K. Urbanowicz , Chad A. Steed , Anton V. Ievlev , Alex Belianinov , Joshua K. Michener , Rama Vasudevan , Olga S. Ovchinnikova
|
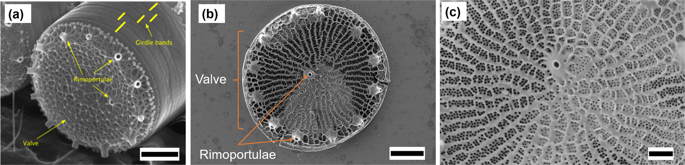
Genome engineering for materials synthesis is a promising avenue for manufacturing materials with unique properties under ambient conditions. Biomineralization in diatoms, unicellular algae that use silica to construct micron-scale cell walls with nanoscale features, is an attractive candidate for functional synthesis of materials for applications including photonics, sensing, filtration, and drug delivery. Therefore, controllably modifying diatom structure through targeted genetic modifications for these applications is a very promising field. In this work, we used gene knockdown in Thalassiosira pseudonana diatoms to create modified strains with changes to structural morphology and linked genotype to phenotype using supervised machine learning. An artificial neural network (NN) was developed to distinguish wild and modified diatoms based on the SEM images of frustules exhibiting phenotypic changes caused by a specific protein (Thaps3_21880), resulting in 94% detection accuracy. Class activation maps visualized physical changes that allowed the NNs to separate diatom strains, subsequently establishing a specific gene that controls pores. A further NN was created to batch process image data, automatically recognize pores, and extract pore-related parameters. Class interrelationship of the extracted paraments was visualized using a multivariate data visualization tool, called CrossVis, and allowed to directly link changes in morphological diatom phenotype of pore size and distribution with changes in the genotype.
中文翻译:

用于硅藻基因工程的深度数据分析,通过机器学习将基因型与表型联系起来
用于材料合成的基因组工程是在环境条件下制造具有独特性能的材料的有前途的途径。硅藻(一种利用二氧化硅构建具有纳米级特征的微米级细胞壁)的单细胞藻类中的生物矿化,是功能性合成材料的有吸引力的候选材料,这些材料可用于光子学,传感,过滤和药物输送。因此,通过针对这些应用的靶向遗传修饰可控地修饰硅藻结构是一个非常有前途的领域。在这项工作中,我们在Thalassiosira中使用了基因敲低利用监督的机器学习技术,利用拟南藻硅藻创建具有结构形态变化的修饰菌株,并将基因型与表型联系起来。开发了一个人工神经网络(NN)来基于显示特定蛋白质(Thaps3_21880)引起的表型变化的壳类的SEM图像来区分野生和修饰的硅藻,从而获得94%的检测精度。类激活图显示了可视化的物理变化,这些变化允许NN分离硅藻菌株,随后建立了控制孔的特定基因。创建了另一个NN,以分批处理图像数据,自动识别孔并提取与孔相关的参数。使用称为CrossVis的多元数据可视化工具可视化提取的参数的类相互关系,











































 京公网安备 11010802027423号
京公网安备 11010802027423号