Human Genome Variation ( IF 1.0 ) Pub Date : 2019-06-07 , DOI: 10.1038/s41439-019-0057-7 Masao Nagasaki , Yoko Kuroki , Tomoko F. Shibata , Fumiki Katsuoka , Takahiro Mimori , Yosuke Kawai , Naoko Minegishi , Atsushi Hozawa , Shinichi Kuriyama , Yoichi Suzuki , Hiroshi Kawame , Fuji Nagami , Takako Takai-Igarashi , Soichi Ogishima , Kaname Kojima , Kazuharu Misawa , Osamu Tanabe , Nobuo Fuse , Hiroshi Tanaka , Nobuo Yaegashi , Kengo Kinoshita , Shiego Kure , Jun Yasuda , Masayuki Yamamoto
|
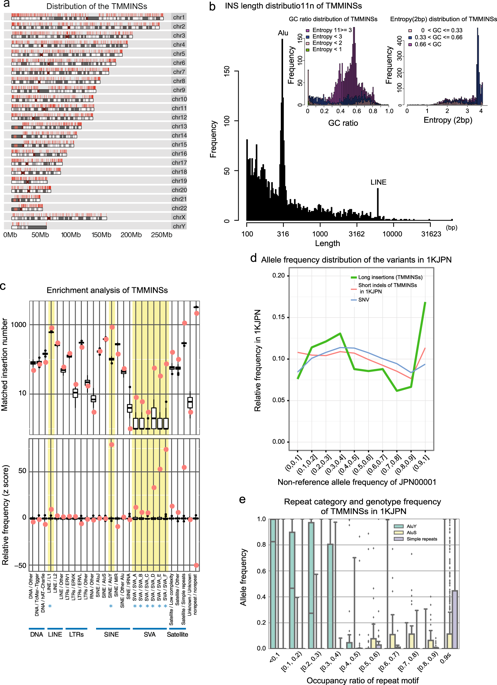
In recent genome analyses, population-specific reference panels have indicated important. However, reference panels based on short-read sequencing data do not sufficiently cover long insertions. Therefore, the nature of long insertions has not been well documented. Here, we assembled a Japanese genome using single-molecule real-time sequencing data and characterized insertions found in the assembled genome. We identified 3691 insertions ranging from 100 bps to ~10,000 bps in the assembled genome relative to the international reference sequence (GRCh38). To validate and characterize these insertions, we mapped short-reads from 1070 Japanese individuals and 728 individuals from eight other populations to insertions integrated into GRCh38. With this result, we constructed JRGv1 (Japanese Reference Genome version 1) by integrating the 903 verified insertions, totaling 1,086,173 bases, shared by at least two Japanese individuals into GRCh38. We also constructed decoyJRGv1 by concatenating 3559 verified insertions, totaling 2,536,870 bases, shared by at least two Japanese individuals or by six other assemblies. This assembly improved the alignment ratio by 0.4% on average. These results demonstrate the importance of refining the reference assembly and creating a population-specific reference genome. JRGv1 and decoyJRGv1 are available at the JRG website.
中文翻译:

利用单分子实时测序技术构建JRG(日本参考基因组)
在最近的基因组分析中,特定于人群的参考指标表明了这一点很重要。但是,基于短读测序数据的参考面板不能充分覆盖长插入片段。因此,长插入的性质尚未得到很好的证明。在这里,我们使用单分子实时测序数据组装了一个日本基因组,并对在组装的基因组中发现的插入进行了表征。相对于国际参考序列(GRCh38),我们在组装的基因组中鉴定了3691个插入序列,范围从100 bps到〜10,000 bps。为了验证和表征这些插入片段,我们将来自1070个日本个体和来自其他八个人群的728个个体的短读段映射到整合到GRCh38中的插入片段。有了这个结果,我们通过整合903个已验证的插入物(共1,086,173个碱基),将至少两个日本人共享的GRCh38片段,构建了JRGv1(日本参考基因组版本1)。我们还通过连接至少3个日本人或其他6个程序集共享的3559个经过验证的插入物(共2536870个碱基)来构建decoyJRGv1。该组件平均将对齐率提高了0.4%。这些结果证明了完善参考装配和创建群体特异性参考基因组的重要性。JRGv1和decoyJRGv1可从JRG网站获得。由至少两个日本人或六个其他议会共享。该组件平均将对齐率提高了0.4%。这些结果证明了完善参考装配和创建群体特异性参考基因组的重要性。JRGv1和decoyJRGv1可从JRG网站获得。由至少两个日本人或六个其他议会共享。该组件平均将对齐比提高了0.4%。这些结果证明了完善参考装配和创建群体特异性参考基因组的重要性。JRGv1和decoyJRGv1可从JRG网站获得。











































 京公网安备 11010802027423号
京公网安备 11010802027423号