当前位置:
X-MOL 学术
›
Prog. Nucl. Magn. Reson. Spectrosc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1ρ relaxation dispersion
Progress in Nuclear Magnetic Resonance Spectroscopy ( IF 7.3 ) Pub Date : 2019-06-01 , DOI: 10.1016/j.pnmrs.2019.05.002 Atul Rangadurai 1 , Eric S Szymaski 1 , Isaac J Kimsey 2 , Honglue Shi 3 , Hashim M Al-Hashimi 4
Progress in Nuclear Magnetic Resonance Spectroscopy ( IF 7.3 ) Pub Date : 2019-06-01 , DOI: 10.1016/j.pnmrs.2019.05.002 Atul Rangadurai 1 , Eric S Szymaski 1 , Isaac J Kimsey 2 , Honglue Shi 3 , Hashim M Al-Hashimi 4
Affiliation
|
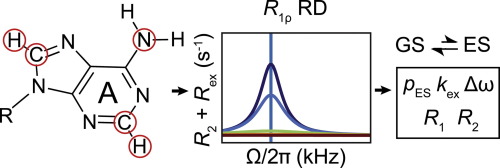
This review describes off-resonance R1ρ relaxation dispersion NMR methods for characterizing microsecond-to-millisecond chemical exchange in uniformly 13C/15N labeled nucleic acids in solution. The review opens with a historical account of key developments that formed the basis for modern R1ρ techniques used to study chemical exchange in biomolecules. A vector model is then used to describe the R1ρ relaxation dispersion experiment, and how the exchange contribution to relaxation varies with the amplitude and frequency offset of an applied spin-locking field, as well as the population, exchange rate, and differences in chemical shifts of two exchanging species. Mathematical treatment of chemical exchange based on the Bloch-McConnell equations is then presented and used to examine relaxation dispersion profiles for more complex exchange scenarios including three-state exchange. Pulse sequences that employ selective Hartmann-Hahn cross-polarization transfers to excite individual 13C or 15N spins are then described for measuring off-resonance R1ρ(13C) and R1ρ(15N) in uniformly 13C/15N labeled DNA and RNA samples prepared using commercially available 13C/15N labeled nucleotide triphosphates. Approaches for analyzing R1ρ data measured at a single static magnetic field to extract a full set of exchange parameters are then presented that rely on numerical integration of the Bloch-McConnell equations or the use of algebraic expressions. Methods for determining structures of nucleic acid excited states are then reviewed that rely on mutations and chemical modifications to bias conformational equilibria, as well as structure-based approaches to calculate chemical shifts. Applications of the methodology to the study of DNA and RNA conformational dynamics are reviewed and the biological significance of the exchange processes is briefly discussed.
中文翻译:

使用非共振 R1ρ 弛豫色散表征核酸中微到毫秒的化学交换
本综述介绍了非共振 R1ρ 弛豫分散 NMR 方法,用于表征溶液中均匀 13C/15N 标记核酸中微秒至毫秒的化学交换。这篇综述首先回顾了关键发展的历史记录,这些发展构成了用于研究生物分子化学交换的现代 R1ρ 技术的基础。然后使用矢量模型来描述 R1ρ 弛豫色散实验,以及交换对弛豫的贡献如何随所应用的自旋锁定场的幅度和频率偏移以及粒子数、交换率和化学位移差异而变化两个交换物种。然后提出了基于布洛赫-麦康奈尔方程的化学交换的数学处理,并将其用于检查更复杂的交换场景(包括三态交换)的弛豫色散分布。然后描述了采用选择性 Hartmann-Hahn 交叉极化转移来激发单个 13C 或 15N 自旋的脉冲序列,用于测量使用市售制备的均匀 13C/15N 标记的 DNA 和 RNA 样品中的偏共振 R1ρ(13C) 和 R1ρ(15N) 13C/15N 标记的三磷酸核苷酸。然后提出了分析在单个静态磁场下测量的 R1ρ 数据以提取全套交换参数的方法,这些方法依赖于 Bloch-McConnell 方程的数值积分或代数表达式的使用。然后回顾了确定核酸激发态结构的方法,这些方法依赖于突变和化学修饰来偏向构象平衡,以及基于结构的方法来计算化学位移。 回顾了该方法在 DNA 和 RNA 构象动力学研究中的应用,并简要讨论了交换过程的生物学意义。
更新日期:2019-06-01
中文翻译:

使用非共振 R1ρ 弛豫色散表征核酸中微到毫秒的化学交换
本综述介绍了非共振 R1ρ 弛豫分散 NMR 方法,用于表征溶液中均匀 13C/15N 标记核酸中微秒至毫秒的化学交换。这篇综述首先回顾了关键发展的历史记录,这些发展构成了用于研究生物分子化学交换的现代 R1ρ 技术的基础。然后使用矢量模型来描述 R1ρ 弛豫色散实验,以及交换对弛豫的贡献如何随所应用的自旋锁定场的幅度和频率偏移以及粒子数、交换率和化学位移差异而变化两个交换物种。然后提出了基于布洛赫-麦康奈尔方程的化学交换的数学处理,并将其用于检查更复杂的交换场景(包括三态交换)的弛豫色散分布。然后描述了采用选择性 Hartmann-Hahn 交叉极化转移来激发单个 13C 或 15N 自旋的脉冲序列,用于测量使用市售制备的均匀 13C/15N 标记的 DNA 和 RNA 样品中的偏共振 R1ρ(13C) 和 R1ρ(15N) 13C/15N 标记的三磷酸核苷酸。然后提出了分析在单个静态磁场下测量的 R1ρ 数据以提取全套交换参数的方法,这些方法依赖于 Bloch-McConnell 方程的数值积分或代数表达式的使用。然后回顾了确定核酸激发态结构的方法,这些方法依赖于突变和化学修饰来偏向构象平衡,以及基于结构的方法来计算化学位移。 回顾了该方法在 DNA 和 RNA 构象动力学研究中的应用,并简要讨论了交换过程的生物学意义。











































 京公网安备 11010802027423号
京公网安备 11010802027423号