当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Parsimonious Gene Correlation Network Analysis (PGCNA): a tool to define modular gene co-expression for refined molecular stratification in cancer.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-04-11 , DOI: 10.1038/s41540-019-0090-7 Matthew A Care 1, 2 , David R Westhead 2 , Reuben M Tooze 1
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-04-11 , DOI: 10.1038/s41540-019-0090-7 Matthew A Care 1, 2 , David R Westhead 2 , Reuben M Tooze 1
Affiliation
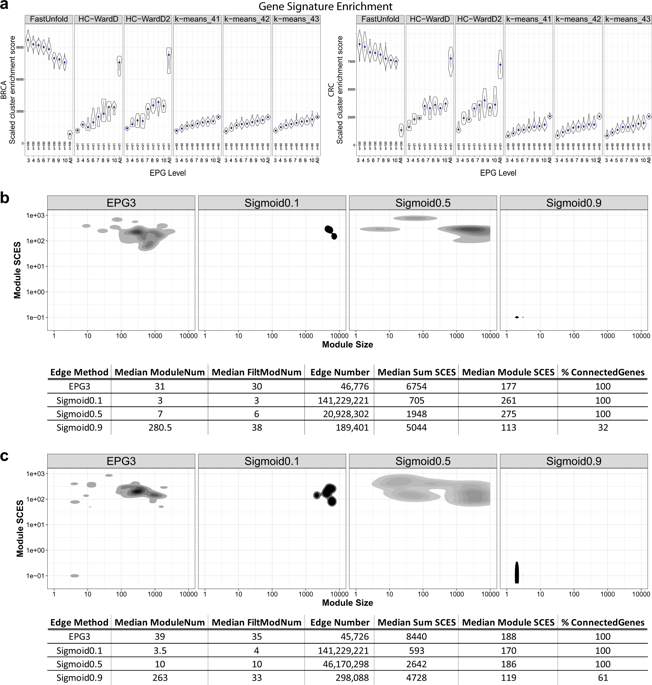
|
Cancers converge onto shared patterns that arise from constraints placed by the biology of the originating cell lineage and microenvironment on programs driven by oncogenic events. Here we define consistent expression modules reflecting this structure in colon and breast cancer by exploiting expression data resources and a new computationally efficient approach that we validate against other comparable methods. This approach, Parsimonious Gene Correlation Network Analysis (PGCNA), allows comparison of network structures between these cancer types identifying shared modules of gene co-expression reflecting: cancer hallmarks, functional and structural gene batteries, copy number variation and biology of originating lineage. These networks along with the mapping of outcome data at gene and module level provide an interactive resource that generates context for relationships between genes within and between such modules. Assigning module expression values (MEVs) provides a tool to summarize network level gene expression in individual cases illustrating potential utility in classification and allowing analysis of linkage between module expression and mutational state. Exploiting TCGA data thus defines both recurrent patterns of association between module expression and mutation at data-set level, and exemplifies the polarization of mutation patterns with the leading edge of module expression at individual case level. We illustrate the scalable nature of the approach within immune response related modules, which in the context of breast cancer demonstrates the selective association of immune subsets, in particular mast cells, with the underlying mutational pattern. Together our analyses provide evidence for a generalizable framework to enhance molecular stratification in cancer.
中文翻译:

简约基因相关网络分析 (PGCNA):一种定义模块化基因共表达的工具,用于癌症中精细的分子分层。
癌症趋向于共同的模式,这些模式是由原始细胞谱系的生物学和微环境对致癌事件驱动的程序所施加的限制而产生的。在这里,我们通过利用表达数据资源和一种新的计算有效的方法来定义反映结肠癌和乳腺癌中这种结构的一致表达模块,并针对其他类似方法进行验证。这种方法称为简约基因相关网络分析 (PGCNA),可以比较这些癌症类型之间的网络结构,识别基因共表达的共享模块,反映:癌症标志、功能和结构基因组、拷贝数变异和起源谱系的生物学。这些网络以及基因和模块水平上结果数据的映射提供了交互式资源,该资源为这些模块内和之间的基因之间的关系生成上下文。分配模块表达值(MEV)提供了一种工具来总结个体案例中的网络水平基因表达,说明分类的潜在效用并允许分析模块表达和突变状态之间的联系。因此,利用 TCGA 数据在数据集级别定义了模块表达和突变之间关联的循环模式,并举例说明了突变模式与个体案例级别模块表达前沿的极化。我们说明了免疫反应相关模块中该方法的可扩展性,在乳腺癌的背景下,该方法证明了免疫亚群(特别是肥大细胞)与潜在突变模式的选择性关联。 我们的分析共同为增强癌症分子分层的通用框架提供了证据。
更新日期:2019-04-11
中文翻译:

简约基因相关网络分析 (PGCNA):一种定义模块化基因共表达的工具,用于癌症中精细的分子分层。
癌症趋向于共同的模式,这些模式是由原始细胞谱系的生物学和微环境对致癌事件驱动的程序所施加的限制而产生的。在这里,我们通过利用表达数据资源和一种新的计算有效的方法来定义反映结肠癌和乳腺癌中这种结构的一致表达模块,并针对其他类似方法进行验证。这种方法称为简约基因相关网络分析 (PGCNA),可以比较这些癌症类型之间的网络结构,识别基因共表达的共享模块,反映:癌症标志、功能和结构基因组、拷贝数变异和起源谱系的生物学。这些网络以及基因和模块水平上结果数据的映射提供了交互式资源,该资源为这些模块内和之间的基因之间的关系生成上下文。分配模块表达值(MEV)提供了一种工具来总结个体案例中的网络水平基因表达,说明分类的潜在效用并允许分析模块表达和突变状态之间的联系。因此,利用 TCGA 数据在数据集级别定义了模块表达和突变之间关联的循环模式,并举例说明了突变模式与个体案例级别模块表达前沿的极化。我们说明了免疫反应相关模块中该方法的可扩展性,在乳腺癌的背景下,该方法证明了免疫亚群(特别是肥大细胞)与潜在突变模式的选择性关联。 我们的分析共同为增强癌症分子分层的通用框架提供了证据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号