当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Synergy from gene expression and network mining (SynGeNet) method predicts synergistic drug combinations for diverse melanoma genomic subtypes.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-02-26 , DOI: 10.1038/s41540-019-0085-4 Kelly E Regan-Fendt 1 , Jielin Xu 1 , Mallory DiVincenzo 2 , Megan C Duggan 2 , Reena Shakya 3 , Ryejung Na 3 , William E Carson 2 , Philip R O Payne 4 , Fuhai Li 4, 5
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2019-02-26 , DOI: 10.1038/s41540-019-0085-4 Kelly E Regan-Fendt 1 , Jielin Xu 1 , Mallory DiVincenzo 2 , Megan C Duggan 2 , Reena Shakya 3 , Ryejung Na 3 , William E Carson 2 , Philip R O Payne 4 , Fuhai Li 4, 5
Affiliation
|
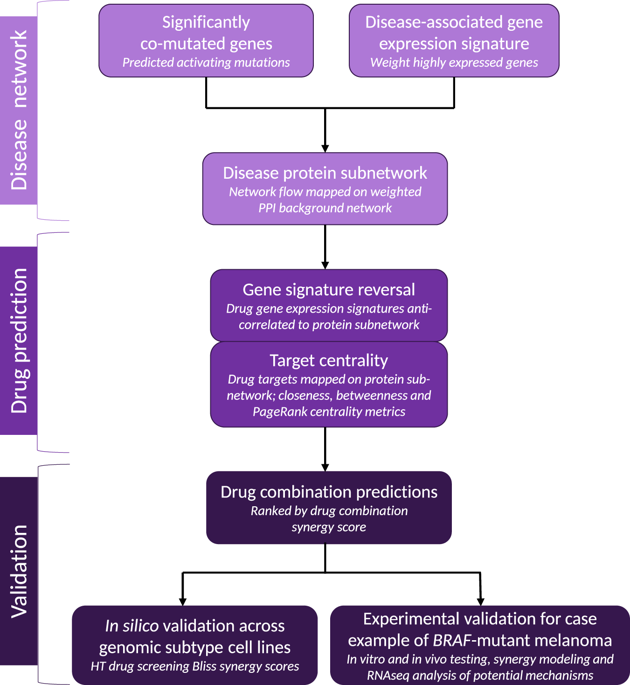
Systems biology perspectives are crucial for understanding the pathophysiology of complex diseases, and therefore hold great promise for the discovery of novel treatment strategies. Drug combinations have been shown to improve durability and reduce resistance to available first-line therapies in a variety of cancers; however, traditional drug discovery approaches are prohibitively cost and labor-intensive to evaluate large-scale matrices of potential drug combinations. Computational methods are needed to efficiently model complex interactions of drug target pathways and identify mechanisms underlying drug combination synergy. In this study, we employ a computational approach, SynGeNet (Synergy from Gene expression and Network mining), which integrates transcriptomics-based connectivity mapping and network centrality analysis to analyze disease networks and predict drug combinations. As an exemplar of a disease in which combination therapies demonstrate efficacy in genomic-specific contexts, we investigate malignant melanoma. We employed SynGeNet to generate drug combination predictions for each of the four major genomic subtypes of melanoma (BRAF, NRAS, NF1, and triple wild type) using publicly available gene expression and mutation data. We validated synergistic drug combinations predicted by our method across all genomic subtypes using results from a high-throughput drug screening study across. Finally, we prospectively validated the drug combination for BRAF-mutant melanoma that was top ranked by our approach, vemurafenib (BRAF inhibitor) + tretinoin (retinoic acid receptor agonist), using both in vitro and in vivo models of BRAF-mutant melanoma and RNA-sequencing analysis of drug-treated melanoma cells to validate the predicted mechanisms. Our approach is applicable to a wide range of disease domains, and, importantly, can model disease-relevant protein subnetworks in precision medicine contexts.
中文翻译:

基因表达和网络挖掘的协同作用 (SynGeNet) 方法可预测不同黑色素瘤基因组亚型的协同药物组合。
系统生物学观点对于理解复杂疾病的病理生理学至关重要,因此为发现新的治疗策略带来了巨大希望。药物组合已被证明可以提高多种癌症的耐久性并减少对现有一线疗法的耐药性;然而,传统的药物发现方法在评估潜在药物组合的大规模矩阵时成本高昂且劳动力密集。需要计算方法来有效地模拟药物靶标途径的复杂相互作用并确定药物组合协同作用的机制。在这项研究中,我们采用了一种计算方法 SynGeNet(基因表达和网络挖掘的协同作用),它集成了基于转录组学的连接图谱和网络中心性分析来分析疾病网络并预测药物组合。作为联合疗法在基因组特异性背景下显示疗效的疾病的一个范例,我们研究了恶性黑色素瘤。我们利用 SynGeNet 使用公开的基因表达和突变数据,对黑色素瘤的四种主要基因组亚型(BRAF、NRAS、NF1 和三重野生型)中的每一种生成药物组合预测。我们使用高通量药物筛选研究的结果验证了我们的方法在所有基因组亚型中预测的协同药物组合。最后,我们使用 BRAF 突变黑色素瘤和 RNA 的体外和体内模型,前瞻性地验证了我们的方法中排名最高的治疗 BRAF 突变黑色素瘤的药物组合,即维莫非尼(BRAF 抑制剂)+ 维甲酸(视黄酸受体激动剂)。 -对药物处理的黑色素瘤细胞进行测序分析,以验证预测的机制。 我们的方法适用于广泛的疾病领域,而且重要的是,可以在精准医学背景下对疾病相关的蛋白质子网络进行建模。
更新日期:2019-02-26
中文翻译:

基因表达和网络挖掘的协同作用 (SynGeNet) 方法可预测不同黑色素瘤基因组亚型的协同药物组合。
系统生物学观点对于理解复杂疾病的病理生理学至关重要,因此为发现新的治疗策略带来了巨大希望。药物组合已被证明可以提高多种癌症的耐久性并减少对现有一线疗法的耐药性;然而,传统的药物发现方法在评估潜在药物组合的大规模矩阵时成本高昂且劳动力密集。需要计算方法来有效地模拟药物靶标途径的复杂相互作用并确定药物组合协同作用的机制。在这项研究中,我们采用了一种计算方法 SynGeNet(基因表达和网络挖掘的协同作用),它集成了基于转录组学的连接图谱和网络中心性分析来分析疾病网络并预测药物组合。作为联合疗法在基因组特异性背景下显示疗效的疾病的一个范例,我们研究了恶性黑色素瘤。我们利用 SynGeNet 使用公开的基因表达和突变数据,对黑色素瘤的四种主要基因组亚型(BRAF、NRAS、NF1 和三重野生型)中的每一种生成药物组合预测。我们使用高通量药物筛选研究的结果验证了我们的方法在所有基因组亚型中预测的协同药物组合。最后,我们使用 BRAF 突变黑色素瘤和 RNA 的体外和体内模型,前瞻性地验证了我们的方法中排名最高的治疗 BRAF 突变黑色素瘤的药物组合,即维莫非尼(BRAF 抑制剂)+ 维甲酸(视黄酸受体激动剂)。 -对药物处理的黑色素瘤细胞进行测序分析,以验证预测的机制。 我们的方法适用于广泛的疾病领域,而且重要的是,可以在精准医学背景下对疾病相关的蛋白质子网络进行建模。











































 京公网安备 11010802027423号
京公网安备 11010802027423号