当前位置:
X-MOL 学术
›
npj Biofilms Microbiomes
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Bacterial release from pipe biofilm in a full-scale drinking water distribution system.
npj Biofilms and Microbiomes ( IF 7.8 ) Pub Date : 2019-02-22 , DOI: 10.1038/s41522-019-0082-9 Sandy Chan 1, 2, 3 , Kristjan Pullerits 1, 2, 3 , Alexander Keucken 4, 5 , Kenneth M Persson 2, 3, 4 , Catherine J Paul 1, 4 , Peter Rådström 1
npj Biofilms and Microbiomes ( IF 7.8 ) Pub Date : 2019-02-22 , DOI: 10.1038/s41522-019-0082-9 Sandy Chan 1, 2, 3 , Kristjan Pullerits 1, 2, 3 , Alexander Keucken 4, 5 , Kenneth M Persson 2, 3, 4 , Catherine J Paul 1, 4 , Peter Rådström 1
Affiliation
|
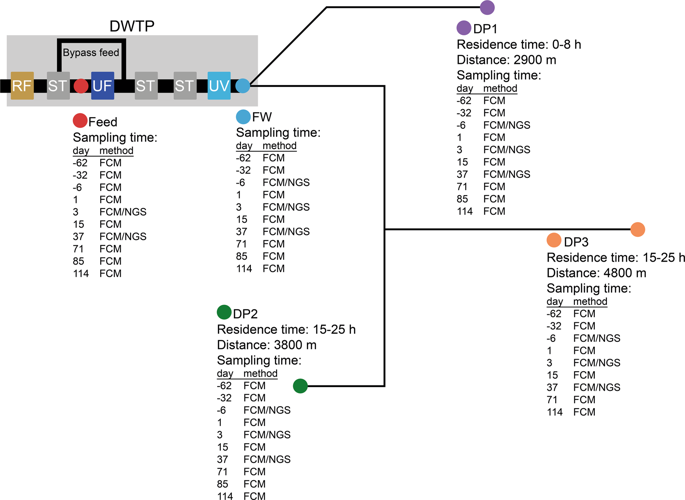
Safe drinking water is delivered to the consumer through kilometres of pipes. These pipes are lined with biofilm, which is thought to affect water quality by releasing bacteria into the drinking water. This study describes the number of cells released from this biofilm, their cellular characteristics, and their identity as they shaped a drinking water microbiome. Installation of ultrafiltration (UF) at full scale in Varberg, Sweden reduced the total cell count to 1.5 × 103 ± 0.5 × 103 cells mL-1 in water leaving the treatment plant. This removed a limitation of both flow cytometry and 16S rRNA amplicon sequencing, which have difficulties in resolving small changes against a high background cell count. Following installation, 58% of the bacteria in the distributed water originated from the pipe biofilm, in contrast to before, when 99.5% of the cells originated from the treatment plant, showing that UF shifts the origin of the drinking water microbiome. The number of bacteria released from the biofilm into the distributed water was 2.1 × 103 ± 1.3 × 103 cells mL-1 and the percentage of HNA (high nucleic acid) content bacteria and intact cells increased as it moved through the distribution system. DESeq2 analysis of 16S rRNA amplicon reads showed increases in 29 operational taxonomic units (OTUs), including genera identified as Sphingomonas, Nitrospira, Mycobacterium, and Hyphomicrobium. This study demonstrated that, due to the installation of UF, the bacteria entering a drinking water microbiome from a pipe biofilm could be both quantitated and described.
中文翻译:

在完整的饮用水分配系统中,管道生物膜中的细菌释放。
安全的饮用水通过数公里的管道输送给消费者。这些管道衬有生物膜,生物膜被认为会通过将细菌释放到饮用水中而影响水质。这项研究描述了从该生物膜释放的细胞数量,它们的细胞特性以及在塑造饮用水微生物组时的身份。在瑞典Varberg大规模安装超滤(UF)后,离开处理厂的水中的总细胞数减少到1.5×103±0.5×103个细胞mL-1。这消除了流式细胞术和16S rRNA扩增子测序的局限性,这些局限性难以解决针对高背景细胞计数的微小变化。安装后,分布水中的细菌中有58%来自管道生物膜,而之前是99。5%的细胞来自处理厂,表明超滤改变了饮用水微生物组的来源。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和次杆菌的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。
更新日期:2019-02-22
中文翻译:

在完整的饮用水分配系统中,管道生物膜中的细菌释放。
安全的饮用水通过数公里的管道输送给消费者。这些管道衬有生物膜,生物膜被认为会通过将细菌释放到饮用水中而影响水质。这项研究描述了从该生物膜释放的细胞数量,它们的细胞特性以及在塑造饮用水微生物组时的身份。在瑞典Varberg大规模安装超滤(UF)后,离开处理厂的水中的总细胞数减少到1.5×103±0.5×103个细胞mL-1。这消除了流式细胞术和16S rRNA扩增子测序的局限性,这些局限性难以解决针对高背景细胞计数的微小变化。安装后,分布水中的细菌中有58%来自管道生物膜,而之前是99。5%的细胞来自处理厂,表明超滤改变了饮用水微生物组的来源。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。从生物膜释放到分布水中的细菌数量为2.1×103±1.3×103细胞mL-1,随着HNA(高核酸)含量的细菌和完整细胞的比例随着分布系统的移动而增加。DESeq2对16S rRNA扩增子读数的分析显示29种操作分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和次杆菌的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。DESeq2对16S rRNA扩增子读数的分析显示29种操作生物分类单位(OTU)有所增加,包括被鉴定为鞘氨醇单胞菌,硝化螺旋菌,分枝杆菌和Hyphomicrobium的属。这项研究表明,由于安装了超滤,可以定量和描述从管道生物膜进入饮用水微生物组的细菌。










































 京公网安备 11010802027423号
京公网安备 11010802027423号