当前位置:
X-MOL 学术
›
Angew. Chem. Int. Ed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Atomic Resolution Insight into Sac7d Protein Binding to DNA and Associated Global Changes by Molecular Dynamics Simulations.
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2019-03-26 , DOI: 10.1002/anie.201900935 Martin Zacharias 1
Angewandte Chemie International Edition ( IF 16.1 ) Pub Date : 2019-03-26 , DOI: 10.1002/anie.201900935 Martin Zacharias 1
Affiliation
|
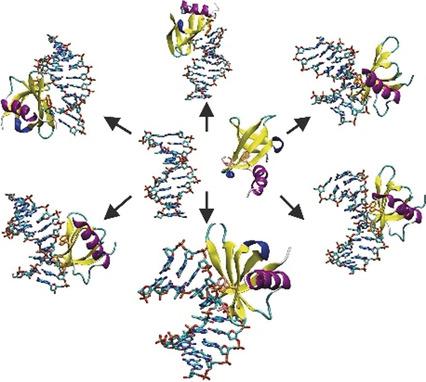
Sac7d is a small, thermostable protein that induces large helical deformations in DNA upon association. Starting from multiple initial placements of the unbound Sac7d structure relative to a B‐DNA oligonucleotide, molecular dynamics (MD) simulations were employed to directly follow several successful binding events at atomic resolution that resulted in structures in close agreement with the native complex geometry. The final native complex formed rapidly within tenths of nanoseconds and included simultaneous large‐scale kinking, groove opening, twisting, and intercalation in the target DNA. The simulations indicate that the complex formation process involves initial non‐native contacts that helped in reaching the final bound state, with residues intercalated at the center of the kinked DNA. It was also possible to identify several long‐lived trapped intermediate states of the binding process and to follow sliding processes of Sac7d along the DNA minor groove.
中文翻译:

通过分子动力学模拟深入了解Sac7d蛋白与DNA的结合以及相关的整体变化。
Sac7d是一种小的热稳定蛋白,在缔合后会引起DNA的大螺旋变形。从相对于B‐DNA寡核苷酸的未结合Sac7d结构的多个初始位置开始,采用分子动力学(MD)模拟以原子分辨率直接追踪多个成功的结合事件,从而导致结构与天然复杂几何形状紧密一致。最终的天然复合物在十分之一纳秒内迅速形成,并且包括目标DNA中的同时大规模扭结,开槽,扭曲和嵌入。模拟表明,复杂的形成过程涉及初始的非天然接触,有助于达到最终的结合状态,残基插入到纽结的DNA中心。
更新日期:2019-03-26
中文翻译:

通过分子动力学模拟深入了解Sac7d蛋白与DNA的结合以及相关的整体变化。
Sac7d是一种小的热稳定蛋白,在缔合后会引起DNA的大螺旋变形。从相对于B‐DNA寡核苷酸的未结合Sac7d结构的多个初始位置开始,采用分子动力学(MD)模拟以原子分辨率直接追踪多个成功的结合事件,从而导致结构与天然复杂几何形状紧密一致。最终的天然复合物在十分之一纳秒内迅速形成,并且包括目标DNA中的同时大规模扭结,开槽,扭曲和嵌入。模拟表明,复杂的形成过程涉及初始的非天然接触,有助于达到最终的结合状态,残基插入到纽结的DNA中心。











































 京公网安备 11010802027423号
京公网安备 11010802027423号