当前位置:
X-MOL 学术
›
Genet. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
CRISPR/Cas9-targeted enrichment and long-read sequencing of the Fuchs endothelial corneal dystrophy-associated TCF4 triplet repeat.
Genetics in Medicine ( IF 6.6 ) Pub Date : 2019-02-08 , DOI: 10.1038/s41436-019-0453-x Nathaniel J Hafford-Tear 1 , Yu-Chih Tsai 2 , Amanda N Sadan 1 , Beatriz Sanchez-Pintado 1 , Christina Zarouchlioti 1 , Geoffrey J Maher 3 , Petra Liskova 1, 4 , Stephen J Tuft 1, 5 , Alison J Hardcastle 1 , Tyson A Clark 2 , Alice E Davidson 1
Genetics in Medicine ( IF 6.6 ) Pub Date : 2019-02-08 , DOI: 10.1038/s41436-019-0453-x Nathaniel J Hafford-Tear 1 , Yu-Chih Tsai 2 , Amanda N Sadan 1 , Beatriz Sanchez-Pintado 1 , Christina Zarouchlioti 1 , Geoffrey J Maher 3 , Petra Liskova 1, 4 , Stephen J Tuft 1, 5 , Alison J Hardcastle 1 , Tyson A Clark 2 , Alice E Davidson 1
Affiliation
|
PURPOSE
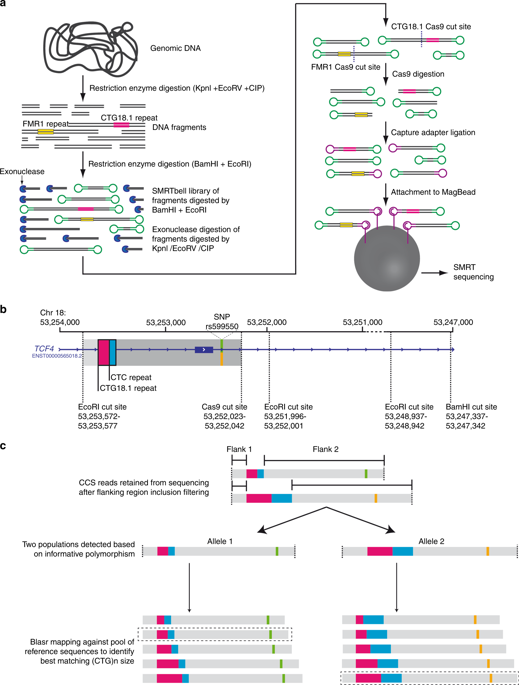
To demonstrate the utility of an amplification-free long-read sequencing method to characterize the Fuchs endothelial corneal dystrophy (FECD)-associated intronic TCF4 triplet repeat (CTG18.1).
METHODS
We applied an amplification-free method, utilizing the CRISPR/Cas9 system, in combination with PacBio single-molecule real-time (SMRT) long-read sequencing, to study CTG18.1. FECD patient samples displaying a diverse range of CTG18.1 allele lengths and zygosity status (n = 11) were analyzed. A robust data analysis pipeline was developed to effectively filter, align, and interrogate CTG18.1-specific reads. All results were compared with conventional polymerase chain reaction (PCR)-based fragment analysis.
RESULTS
CRISPR-guided SMRT sequencing of CTG18.1 provided accurate genotyping information for all samples and phasing was possible for 18/22 alleles sequenced. Repeat length instability was observed for all expanded (≥50 repeats) phased CTG18.1 alleles analyzed. Furthermore, higher levels of repeat instability were associated with increased CTG18.1 allele length (mode length ≥91 repeats) indicating that expanded alleles behave dynamically.
CONCLUSION
CRISPR-guided SMRT sequencing of CTG18.1 has revealed novel insights into CTG18.1 length instability. Furthermore, this study provides a framework to improve the molecular diagnostic accuracy for CTG18.1-mediated FECD, which we anticipate will become increasingly important as gene-directed therapies are developed for this common age-related and sight threatening disease.
中文翻译:

对 Fuchs 内皮性角膜营养不良相关的 TCF4 三联体重复序列进行 CRISPR/Cas9 靶向富集和长读长测序。
目的 证明无扩增长读长测序方法在表征 Fuchs 内皮性角膜营养不良 (FECD) 相关内含子 TCF4 三联体重复序列 (CTG18.1) 的效用。方法我们采用免扩增方法,利用CRISPR/Cas9系统,结合PacBio单分子实时(SMRT)长读长测序来研究CTG18.1。对显示不同范围的 CTG18.1 等位基因长度和接合状态 (n = 11) 的 FECD 患者样本进行了分析。开发了强大的数据分析管道来有效过滤、对齐和询问 CTG18.1 特定的读数。所有结果均与基于传统聚合酶链式反应 (PCR) 的片段分析进行比较。结果 CTG18.1 的 CRISPR 引导 SMRT 测序为所有样本提供了准确的基因分型信息,并且可以对已测序的 18/22 等位基因进行定相。对于分析的所有扩展(≥50 次重复)定相 CTG18.1 等位基因,观察到重复长度不稳定。此外,较高水平的重复不稳定性与 CTG18.1 等位基因长度增加(众数长度≥91 次重复)相关,表明扩展的等位基因表现动态。结论 CTG18.1 的 CRISPR 引导 SMRT 测序揭示了对 CTG18.1 长度不稳定性的新见解。此外,这项研究提供了一个框架来提高 CTG18.1 介导的 FECD 的分子诊断准确性,随着针对这种常见的年龄相关和视力威胁疾病的基因定向疗法的开发,我们预计该框架将变得越来越重要。
更新日期:2019-02-08
中文翻译:

对 Fuchs 内皮性角膜营养不良相关的 TCF4 三联体重复序列进行 CRISPR/Cas9 靶向富集和长读长测序。
目的 证明无扩增长读长测序方法在表征 Fuchs 内皮性角膜营养不良 (FECD) 相关内含子 TCF4 三联体重复序列 (CTG18.1) 的效用。方法我们采用免扩增方法,利用CRISPR/Cas9系统,结合PacBio单分子实时(SMRT)长读长测序来研究CTG18.1。对显示不同范围的 CTG18.1 等位基因长度和接合状态 (n = 11) 的 FECD 患者样本进行了分析。开发了强大的数据分析管道来有效过滤、对齐和询问 CTG18.1 特定的读数。所有结果均与基于传统聚合酶链式反应 (PCR) 的片段分析进行比较。结果 CTG18.1 的 CRISPR 引导 SMRT 测序为所有样本提供了准确的基因分型信息,并且可以对已测序的 18/22 等位基因进行定相。对于分析的所有扩展(≥50 次重复)定相 CTG18.1 等位基因,观察到重复长度不稳定。此外,较高水平的重复不稳定性与 CTG18.1 等位基因长度增加(众数长度≥91 次重复)相关,表明扩展的等位基因表现动态。结论 CTG18.1 的 CRISPR 引导 SMRT 测序揭示了对 CTG18.1 长度不稳定性的新见解。此外,这项研究提供了一个框架来提高 CTG18.1 介导的 FECD 的分子诊断准确性,随着针对这种常见的年龄相关和视力威胁疾病的基因定向疗法的开发,我们预计该框架将变得越来越重要。











































 京公网安备 11010802027423号
京公网安备 11010802027423号