当前位置:
X-MOL 学术
›
Mol. Biosyst.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of a suitable promoter for the sigma factor of Mycobacterium tuberculosis
Molecular BioSystems Pub Date : 2017-09-12 00:00:00 , DOI: 10.1039/c7mb00317j A. Mallick Gupta 1, 2, 3, 4 , S. Mukherjee 3, 4, 5, 6 , A. Dutta 3, 4, 7 , J. Mukhopadhyay 3, 4, 7 , D. Bhattacharyya 3, 4, 5, 6 , S. Mandal 1, 2, 3, 4
Molecular BioSystems Pub Date : 2017-09-12 00:00:00 , DOI: 10.1039/c7mb00317j A. Mallick Gupta 1, 2, 3, 4 , S. Mukherjee 3, 4, 5, 6 , A. Dutta 3, 4, 7 , J. Mukhopadhyay 3, 4, 7 , D. Bhattacharyya 3, 4, 5, 6 , S. Mandal 1, 2, 3, 4
Affiliation
|
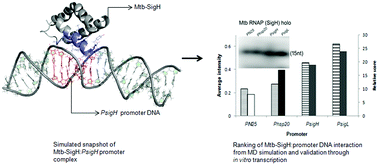
Promoter binding specificity is one of the important characteristics of transcription by Mycobacterium tuberculosis (Mtb) sigma (σ) factors, which remains unexplored due to limited structural evidence. Our previous study on the structural features of Mtb-SigH, consisting of three alpha helices, and its interaction with core RNA polymerase has been extended herein to determine the little known DNA sequence recognition pattern involving its cognate promoters. Herein, high resolution X-ray crystallographic structures of the protein–DNA complexes were inspected to determine the tentative DNA-binding helix of the σ factor. The binding interface in the available crystal structures is found to be populated mainly with specific residues such as Arg, Asn, Lys, Gln, and Ser. We uncovered the helix 3 of Mtb-SigH containing most of these amino acids, which ranged from Arg 64 to Arg 75, forming the predicted active site. The complex of Mtb-SigH:DNA is modelled with 20 promoter sequences. The binding affinity is predicted by scoring these protein–DNA complexes through proximity and interaction parameters obtained by molecular dynamics simulations. The promoters are ranked considering hydrogen bonding, energy of interaction, buried surface area, and distance between centers of masses in interaction with the protein. The ranking is validated through in vitro transcription assays. The trends of these selected promoter interactions have shown variations parallel to the experimental evaluation, emphasizing the success of the active site determination along with screening of the promoter strength. The promoter interaction of Mtb-SigH can be highly beneficial for understanding the regulation of gene expression of a pathogen and also extends a solid platform to predict promoters for other bacterial σ factors.
中文翻译:

鉴定结核分枝杆菌sigma因子的合适启动子
启动子结合特异性是结核分枝杆菌转录的重要特征之一。(Mtb)sigma(σ)因子,由于结构证据有限而仍未开发。我们先前对由三个α螺旋组成的Mtb-SigH的结构特征及其与核心RNA聚合酶的相互作用的研究已在本文中扩展,以确定涉及其同源启动子的鲜为人知的DNA序列识别模式。在此,检查了蛋白质-DNA复合物的高分辨率X射线晶体学结构,以确定σ因子的暂定DNA结合螺旋。发现可用晶体结构中的结合界面主要填充有特定残基,例如Arg,Asn,Lys,Gln和Ser。我们发现了Mtb-SigH的螺旋3,其中包含大多数这些氨基酸,其范围从Arg 64到Arg 75,形成了预测的活性位点。Mtb-SigH的复合体:用20个启动子序列对DNA进行建模。通过通过分子动力学模拟获得的邻近性和相互作用参数对这些蛋白质-DNA复合物评分,可以预测结合亲和力。考虑到氢键,相互作用的能量,埋入的表面积以及与蛋白质相互作用的质心之间的距离,对启动子进行排名。排名通过验证体外转录测定。这些选择的启动子相互作用的趋势已经显示出与实验评估平行的变化,强调了活性位点确定以及启动子强度筛选的成功。Mtb-SigH的启动子相互作用对于理解病原体基因表达的调控可能是非常有益的,并且还为预测其他细菌σ因子的启动子提供了坚实的平台。
更新日期:2017-10-25
中文翻译:

鉴定结核分枝杆菌sigma因子的合适启动子
启动子结合特异性是结核分枝杆菌转录的重要特征之一。(Mtb)sigma(σ)因子,由于结构证据有限而仍未开发。我们先前对由三个α螺旋组成的Mtb-SigH的结构特征及其与核心RNA聚合酶的相互作用的研究已在本文中扩展,以确定涉及其同源启动子的鲜为人知的DNA序列识别模式。在此,检查了蛋白质-DNA复合物的高分辨率X射线晶体学结构,以确定σ因子的暂定DNA结合螺旋。发现可用晶体结构中的结合界面主要填充有特定残基,例如Arg,Asn,Lys,Gln和Ser。我们发现了Mtb-SigH的螺旋3,其中包含大多数这些氨基酸,其范围从Arg 64到Arg 75,形成了预测的活性位点。Mtb-SigH的复合体:用20个启动子序列对DNA进行建模。通过通过分子动力学模拟获得的邻近性和相互作用参数对这些蛋白质-DNA复合物评分,可以预测结合亲和力。考虑到氢键,相互作用的能量,埋入的表面积以及与蛋白质相互作用的质心之间的距离,对启动子进行排名。排名通过验证体外转录测定。这些选择的启动子相互作用的趋势已经显示出与实验评估平行的变化,强调了活性位点确定以及启动子强度筛选的成功。Mtb-SigH的启动子相互作用对于理解病原体基因表达的调控可能是非常有益的,并且还为预测其他细菌σ因子的启动子提供了坚实的平台。











































 京公网安备 11010802027423号
京公网安备 11010802027423号