当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Employing unnatural promiscuity of sortase to construct peptide macrocycle libraries for ligand discovery
Chemical Science ( IF 8.4 ) Pub Date : 2024-05-22 , DOI: 10.1039/d4sc01992j Yan-Ni Zhang 1 , Xiao-Cui Wan 1 , Yang Tang 2 , Ying Chen 1 , Feng-Hao Zheng 1 , Zhi-Hui Cui 1 , Hua Zhang 1 , Zhaocai Zhou 3 , Ge-Min Fang 1
Chemical Science ( IF 8.4 ) Pub Date : 2024-05-22 , DOI: 10.1039/d4sc01992j Yan-Ni Zhang 1 , Xiao-Cui Wan 1 , Yang Tang 2 , Ying Chen 1 , Feng-Hao Zheng 1 , Zhi-Hui Cui 1 , Hua Zhang 1 , Zhaocai Zhou 3 , Ge-Min Fang 1
Affiliation
|
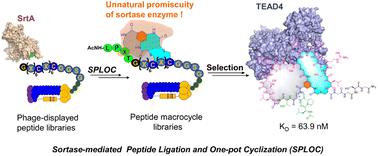
With the increasing attention paid to macrocyclic scaffolds in peptide drug development, genetically encoded peptide macrocycle libraries have become invaluable sources for the discovery of high-affinity peptide ligands targeting disease-associated proteins. The traditional phage display technique of constructing disulfide-tethered macrocycles by cysteine oxidation has the inherent drawback of reduction instability of the disulfide bond. Chemical macrocyclization solves the problem of disulfide bond instability, but the involved highly electrophilic reagents are usually toxic to phages and may bring undesirable side reactions. Here, we report a unique Sortase-mediated Peptide Ligation and One-pot Cyclization strategy (SPLOC) to generate peptide macrocycle libraries, avoiding the undesired reactions of electrophiles with phages. The key to this platform is to mine the unnatural promiscuity of sortase on the X residue of the pentapeptide recognition sequence (LPXTG). Low reactive electrophiles are incorporated into the X-residue side chain, enabling intramolecular cyclization with the cysteine residue of the phage-displayed peptide library. Utilizing the genetically encoded peptide macrocycle library constructed by the SPLOC platform, we found a high-affinity bicyclic peptide binding TEAD4 with a nanomolar KD value (63.9 nM). Importantly, the binding affinity of the bicyclic peptide ligand is 102-fold lower than that of the acyclic analogue. To our knowledge, this is the first time to mine the unnatural promiscuity of ligases to generate peptide macrocycles, providing a new avenue for the construction of genetically encoded cyclic peptide libraries.
中文翻译:

利用分选酶的非自然混杂性构建肽大环文库以发现配体
随着肽药物开发中大环支架的日益受到关注,基因编码的肽大环文库已成为发现针对疾病相关蛋白的高亲和力肽配体的宝贵来源。传统的通过半胱氨酸氧化构建二硫键大环化合物的噬菌体展示技术具有二硫键还原不稳定的固有缺点。化学大环化解决了二硫键不稳定的问题,但所涉及的高亲电试剂通常对噬菌体有毒,并可能带来不良副反应。在这里,我们报告了一种独特的分选酶介导的肽连接和一锅环化策略(SPLOC)来生成肽大环文库,避免亲电子试剂与噬菌体发生不良反应。该平台的关键是挖掘分选酶在五肽识别序列(LPXTG)的X残基上的非自然混杂性。低反应性亲电子试剂被整合到 X 残基侧链中,从而能够与噬菌体展示肽库的半胱氨酸残基进行分子内环化。利用SPLOC平台构建的基因编码肽大环文库,我们发现了一种高亲和力的双环肽结合TEAD4,其KD值为纳摩尔级(63.9 nM)。重要的是,双环肽配体的结合亲和力比无环类似物低 102 倍。据我们所知,这是首次挖掘连接酶的非自然混杂性来生成肽大环,为构建基因编码的环肽库提供了新途径。
更新日期:2024-05-22
中文翻译:

利用分选酶的非自然混杂性构建肽大环文库以发现配体
随着肽药物开发中大环支架的日益受到关注,基因编码的肽大环文库已成为发现针对疾病相关蛋白的高亲和力肽配体的宝贵来源。传统的通过半胱氨酸氧化构建二硫键大环化合物的噬菌体展示技术具有二硫键还原不稳定的固有缺点。化学大环化解决了二硫键不稳定的问题,但所涉及的高亲电试剂通常对噬菌体有毒,并可能带来不良副反应。在这里,我们报告了一种独特的分选酶介导的肽连接和一锅环化策略(SPLOC)来生成肽大环文库,避免亲电子试剂与噬菌体发生不良反应。该平台的关键是挖掘分选酶在五肽识别序列(LPXTG)的X残基上的非自然混杂性。低反应性亲电子试剂被整合到 X 残基侧链中,从而能够与噬菌体展示肽库的半胱氨酸残基进行分子内环化。利用SPLOC平台构建的基因编码肽大环文库,我们发现了一种高亲和力的双环肽结合TEAD4,其KD值为纳摩尔级(63.9 nM)。重要的是,双环肽配体的结合亲和力比无环类似物低 102 倍。据我们所知,这是首次挖掘连接酶的非自然混杂性来生成肽大环,为构建基因编码的环肽库提供了新途径。

































 京公网安备 11010802027423号
京公网安备 11010802027423号