当前位置:
X-MOL 学术
›
Environ. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
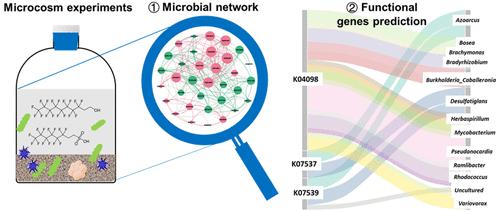
Using Network Analysis and Predictive Functional Analysis to Explore the Fluorotelomer Biotransformation Potential of Soil Microbial Communities
Environmental Science & Technology ( IF 11.4 ) Pub Date : 2024-04-19 , DOI: 10.1021/acs.est.4c00942 Sheng Dong 1 , Peng-Fei Yan 1 , Melissa P. Mezzari 2 , Linda M. Abriola 3 , Kurt D. Pennell 3 , Natalie L. Cápiro 1
Environmental Science & Technology ( IF 11.4 ) Pub Date : 2024-04-19 , DOI: 10.1021/acs.est.4c00942 Sheng Dong 1 , Peng-Fei Yan 1 , Melissa P. Mezzari 2 , Linda M. Abriola 3 , Kurt D. Pennell 3 , Natalie L. Cápiro 1
Affiliation
|
Microbial transformation of per- and polyfluoroalkyl substances (PFAS), including fluorotelomer-derived PFAS, by native microbial communities in the environment has been widely documented. However, few studies have identified the key microorganisms and their roles during the PFAS biotransformation processes. This study was undertaken to gain more insight into the structure and function of soil microbial communities that are relevant to PFAS biotransformation. We collected 16S rRNA gene sequencing data from 8:2 fluorotelomer alcohol and 6:2 fluorotelomer sulfonate biotransformation studies conducted in soil microcosms under various redox conditions. Through co-occurrence network analysis, several genera, including Variovorax, Rhodococcus, and Cupriavidus, were found to likely play important roles in the biotransformation of fluorotelomers. Additionally, a metagenomic prediction approach (PICRUSt2) identified functional genes, including 6-oxocyclohex-1-ene-carbonyl-CoA hydrolase, cyclohexa-1,5-dienecarbonyl-CoA hydratase, and a fluoride-proton antiporter gene, that may be involved in defluorination. This study pioneers the application of these bioinformatics tools in the analysis of PFAS biotransformation-related sequencing data. Our findings serve as a foundational reference for investigating enzymatic mechanisms of microbial defluorination that may facilitate the development of efficient microbial consortia and/or pure microbial strains for PFAS biotransformation.
中文翻译:

利用网络分析和预测功能分析探索土壤微生物群落的氟调聚物生物转化潜力
环境中原生微生物群落对全氟烷基物质和多氟烷基物质 (PFAS)(包括氟调聚物衍生的 PFAS)的微生物转化已被广泛记录。然而,很少有研究确定关键微生物及其在 PFAS 生物转化过程中的作用。本研究旨在更深入地了解与 PFAS 生物转化相关的土壤微生物群落的结构和功能。我们收集了在各种氧化还原条件下在土壤微观世界中进行的 8:2 氟调聚物醇和 6:2 氟调聚物磺酸盐生物转化研究的 16S rRNA 基因测序数据。通过共现网络分析,发现包括Variovorax、红球菌属和Cupriavidus在内的几个属可能在含氟调聚物的生物转化中发挥重要作用。此外,宏基因组预测方法 (PICRUSt2) 鉴定了可能涉及的功能基因,包括 6-氧代环己-1-烯-羰基-CoA 水解酶、环己-1,5-二烯羰基-CoA 水合酶和氟化物-质子逆向转运蛋白基因。在除氟中。这项研究开创了这些生物信息学工具在 PFAS 生物转化相关测序数据分析中的应用。我们的研究结果为研究微生物脱氟的酶促机制提供了基础参考,这可能有助于开发用于 PFAS 生物转化的有效微生物群落和/或纯微生物菌株。
更新日期:2024-04-19
中文翻译:

利用网络分析和预测功能分析探索土壤微生物群落的氟调聚物生物转化潜力
环境中原生微生物群落对全氟烷基物质和多氟烷基物质 (PFAS)(包括氟调聚物衍生的 PFAS)的微生物转化已被广泛记录。然而,很少有研究确定关键微生物及其在 PFAS 生物转化过程中的作用。本研究旨在更深入地了解与 PFAS 生物转化相关的土壤微生物群落的结构和功能。我们收集了在各种氧化还原条件下在土壤微观世界中进行的 8:2 氟调聚物醇和 6:2 氟调聚物磺酸盐生物转化研究的 16S rRNA 基因测序数据。通过共现网络分析,发现包括Variovorax、红球菌属和Cupriavidus在内的几个属可能在含氟调聚物的生物转化中发挥重要作用。此外,宏基因组预测方法 (PICRUSt2) 鉴定了可能涉及的功能基因,包括 6-氧代环己-1-烯-羰基-CoA 水解酶、环己-1,5-二烯羰基-CoA 水合酶和氟化物-质子逆向转运蛋白基因。在除氟中。这项研究开创了这些生物信息学工具在 PFAS 生物转化相关测序数据分析中的应用。我们的研究结果为研究微生物脱氟的酶促机制提供了基础参考,这可能有助于开发用于 PFAS 生物转化的有效微生物群落和/或纯微生物菌株。



























 京公网安备 11010802027423号
京公网安备 11010802027423号