当前位置:
X-MOL 学术
›
J. Biomed. Inform.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
A comprehensive performance evaluation, comparison, and integration of computational methods for detecting and estimating cross-contamination of human samples in cancer next-generation sequencing analysis
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2024-03-12 , DOI: 10.1016/j.jbi.2024.104625 Huijuan Chen , Bing Wang , Lili Cai , Xiaotian Yang , Yali Hu , Yiran Zhang , Xue Leng , Wen Liu , Dongjie Fan , Beifang Niu , Qiming Zhou
Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2024-03-12 , DOI: 10.1016/j.jbi.2024.104625 Huijuan Chen , Bing Wang , Lili Cai , Xiaotian Yang , Yali Hu , Yiran Zhang , Xue Leng , Wen Liu , Dongjie Fan , Beifang Niu , Qiming Zhou
|
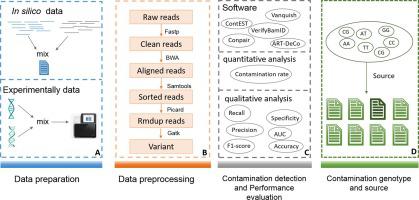
Cross-sample contamination is one of the major issues in next-generation sequencing (NGS)-based molecular assays. This type of contamination, even at very low levels, can significantly impact the results of an analysis, especially in the detection of somatic alterations in tumor samples. Several contamination identification tools have been developed and implemented as a crucial quality-control step in the routine NGS bioinformatic pipeline. However, no study has been published to comprehensively and systematically investigate, evaluate, and compare these computational methods in the cancer NGS analysis. In this study, we comprehensively investigated nine state-of-the-art computational methods for detecting cross-sample contamination. To explore their application in cancer NGS analysis, we further compared the performance of five representative tools by qualitative and quantitative analyses using and simulated experimental NGS data. The results showed that Conpair achieved the best performance for identifying contamination and predicting the level of contamination in solid tumors NGS analysis. Moreover, based on Conpair, we developed a Python script, Contamination Source Predictor (ConSPr), to identify the source of contamination. We anticipate that this comprehensive survey and the proposed tool for predicting the source of contamination will assist researchers in selecting appropriate cross-contamination detection tools in cancer NGS analysis and inspire the development of computational methods for detecting sample cross-contamination and identifying its source in the future.
中文翻译:

用于检测和估计癌症下一代测序分析中人类样本交叉污染的计算方法的综合性能评估、比较和集成
交叉样本污染是基于下一代测序 (NGS) 的分子检测中的主要问题之一。这种类型的污染,即使水平非常低,也会显着影响分析结果,特别是在检测肿瘤样本中的体细胞变化时。已经开发并实施了多种污染识别工具,作为常规 NGS 生物信息学流程中的关键质量控制步骤。然而,目前还没有发表研究来全面、系统地研究、评估和比较这些计算方法在癌症 NGS 分析中的应用。在这项研究中,我们全面研究了九种用于检测交叉样本污染的最先进的计算方法。为了探索它们在癌症 NGS 分析中的应用,我们使用模拟实验 NGS 数据通过定性和定量分析进一步比较了五种代表性工具的性能。结果表明,Conpair 在实体瘤 NGS 分析中识别污染和预测污染水平方面取得了最佳性能。此外,基于Conpair,我们开发了Python脚本Contamination Source Predictor(ConSPr)来识别污染源。我们预计,这项全面的调查和提出的预测污染源的工具将有助于研究人员在癌症 NGS 分析中选择合适的交叉污染检测工具,并激发用于检测样本交叉污染并识别其来源的计算方法的开发。未来。
更新日期:2024-03-12
中文翻译:

用于检测和估计癌症下一代测序分析中人类样本交叉污染的计算方法的综合性能评估、比较和集成
交叉样本污染是基于下一代测序 (NGS) 的分子检测中的主要问题之一。这种类型的污染,即使水平非常低,也会显着影响分析结果,特别是在检测肿瘤样本中的体细胞变化时。已经开发并实施了多种污染识别工具,作为常规 NGS 生物信息学流程中的关键质量控制步骤。然而,目前还没有发表研究来全面、系统地研究、评估和比较这些计算方法在癌症 NGS 分析中的应用。在这项研究中,我们全面研究了九种用于检测交叉样本污染的最先进的计算方法。为了探索它们在癌症 NGS 分析中的应用,我们使用模拟实验 NGS 数据通过定性和定量分析进一步比较了五种代表性工具的性能。结果表明,Conpair 在实体瘤 NGS 分析中识别污染和预测污染水平方面取得了最佳性能。此外,基于Conpair,我们开发了Python脚本Contamination Source Predictor(ConSPr)来识别污染源。我们预计,这项全面的调查和提出的预测污染源的工具将有助于研究人员在癌症 NGS 分析中选择合适的交叉污染检测工具,并激发用于检测样本交叉污染并识别其来源的计算方法的开发。未来。



























 京公网安备 11010802027423号
京公网安备 11010802027423号