当前位置:
X-MOL 学术
›
Comput. Biol. Med.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Transcriptomic signature of cancer cachexia by integration of machine learning, literature mining and meta-analysis
Computers in Biology and Medicine ( IF 7.7 ) Pub Date : 2024-02-28 , DOI: 10.1016/j.compbiomed.2024.108233 Kening Zhao , Esmaeil Ebrahimie , Manijeh Mohammadi Dehcheshmeh , Mathew G. Lewsey , Lei Zheng , Nick J. Hoogenraad
Computers in Biology and Medicine ( IF 7.7 ) Pub Date : 2024-02-28 , DOI: 10.1016/j.compbiomed.2024.108233 Kening Zhao , Esmaeil Ebrahimie , Manijeh Mohammadi Dehcheshmeh , Mathew G. Lewsey , Lei Zheng , Nick J. Hoogenraad
|
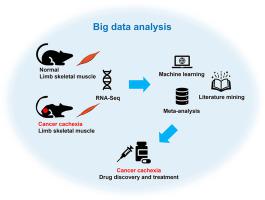
Cancer cachexia is a severe metabolic syndrome marked by skeletal muscle atrophy. A successful clinical intervention for cancer cachexia is currently lacking. The study of cachexia mechanisms is largely based on preclinical animal models and the availability of high-throughput transcriptomic datasets of cachectic mouse muscles is increasing through the extensive use of next generation sequencing technologies. Cachectic mouse muscle transcriptomic datasets of ten different studies were combined and mined by seven attribute weighting models, which analysed both categorical variables and numerical variables. The transcriptomic signature of cancer cachexia was identified by attribute weighting algorithms and was used to evaluate the performance of eleven pattern discovery models. The signature was employed to find the best combination of drugs (drug repurposing) for developing cancer cachexia treatment strategies, as well as to evaluate currently used cachexia drugs by literature mining. Attribute weighting algorithms ranked 26 genes as the transcriptomic signature of muscle from mice with cancer cachexia. Deep Learning and Random Forest models performed better in differentiating cancer cachexia cases based on muscle transcriptomic data. Literature mining revealed that a combination of melatonin and infliximab has negative interactions with 2 key genes ( and ) upregulated in the transcriptomic signature of cancer cachexia in muscle. The integration of machine learning, meta-analysis and literature mining was found to be an efficient approach to identifying a robust transcriptomic signature for cancer cachexia, with implications for improving clinical diagnosis and management of this condition.
中文翻译:

机器学习、文献挖掘和荟萃分析相结合的癌症恶病质转录组学特征
癌症恶病质是一种以骨骼肌萎缩为特征的严重代谢综合征。目前缺乏针对癌症恶病质的成功临床干预。恶病质机制的研究主要基于临床前动物模型,并且通过下一代测序技术的广泛使用,恶病质小鼠肌肉的高通量转录组数据集的可用性正在增加。十项不同研究的恶病质小鼠肌肉转录组数据集通过七个属性加权模型进行组合和挖掘,该模型分析了分类变量和数值变量。通过属性加权算法识别癌症恶病质的转录组特征,并用于评估十一种模式发现模型的性能。该签名用于寻找最佳药物组合(药物再利用)来制定癌症恶病质治疗策略,并通过文献挖掘评估当前使用的恶病质药物。属性加权算法将 26 个基因排名为癌症恶病质小鼠肌肉的转录组特征。深度学习和随机森林模型在基于肌肉转录组数据区分癌症恶病质病例方面表现更好。文献挖掘表明,褪黑素和英夫利昔单抗的组合与肌肉癌症恶病质转录组特征上调的 2 个关键基因 ( 和 ) 存在负相互作用。人们发现,机器学习、荟萃分析和文献挖掘的整合是识别癌症恶病质的强大转录组学特征的有效方法,对于改善这种情况的临床诊断和管理具有重要意义。
更新日期:2024-02-28
中文翻译:

机器学习、文献挖掘和荟萃分析相结合的癌症恶病质转录组学特征
癌症恶病质是一种以骨骼肌萎缩为特征的严重代谢综合征。目前缺乏针对癌症恶病质的成功临床干预。恶病质机制的研究主要基于临床前动物模型,并且通过下一代测序技术的广泛使用,恶病质小鼠肌肉的高通量转录组数据集的可用性正在增加。十项不同研究的恶病质小鼠肌肉转录组数据集通过七个属性加权模型进行组合和挖掘,该模型分析了分类变量和数值变量。通过属性加权算法识别癌症恶病质的转录组特征,并用于评估十一种模式发现模型的性能。该签名用于寻找最佳药物组合(药物再利用)来制定癌症恶病质治疗策略,并通过文献挖掘评估当前使用的恶病质药物。属性加权算法将 26 个基因排名为癌症恶病质小鼠肌肉的转录组特征。深度学习和随机森林模型在基于肌肉转录组数据区分癌症恶病质病例方面表现更好。文献挖掘表明,褪黑素和英夫利昔单抗的组合与肌肉癌症恶病质转录组特征上调的 2 个关键基因 ( 和 ) 存在负相互作用。人们发现,机器学习、荟萃分析和文献挖掘的整合是识别癌症恶病质的强大转录组学特征的有效方法,对于改善这种情况的临床诊断和管理具有重要意义。



























 京公网安备 11010802027423号
京公网安备 11010802027423号