Journal of Biomedical informatics ( IF 4.5 ) Pub Date : 2023-12-15 , DOI: 10.1016/j.jbi.2023.104569 Federica Cruciani , Antonino Aparo , Lorenza Brusini , Carlo Combi , Silvia F. Storti , Rosalba Giugno , Gloria Menegaz , Ilaria Boscolo Galazzo
|
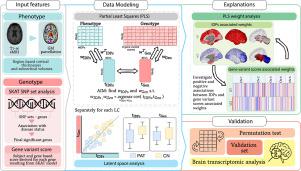
The joint modeling of genetic data and brain imaging information allows for determining the pathophysiological pathways of neurodegenerative diseases such as Alzheimer’s disease (AD). This task has typically been approached using mass-univariate methods that rely on a complete set of Single Nucleotide Polymorphisms (SNPs) to assess their association with selected image-derived phenotypes (IDPs). However, such methods are prone to multiple comparisons bias and, most importantly, fail to account for potential cross-feature interactions, resulting in insufficient detection of significant associations. Ways to overcome these limitations while reducing the number of traits aim at conveying genetic information at the gene level and capturing the integrated genetic effects of a set of genetic variants, rather than looking at each SNP individually. Their associations with brain IDPs are still largely unexplored in the current literature, though they can uncover new potential genetic determinants for brain modulations in the AD continuum. In this work, we explored an explainable multivariate model to analyze the genetic basis of the grey matter modulations, relying on the AD Neuroimaging Initiative (ADNI) phase 3 dataset. Cortical thicknesses and subcortical volumes derived from T1-weighted Magnetic Resonance were considered to describe the imaging phenotypes. At the same time the genetic counterpart was represented by gene variant scores extracted by the Sequence Kernel Association Test (SKAT) filtering model. Moreover, transcriptomic analysis was carried on to assess the expression of the resulting genes in the main brain structures as a form of validation. Results highlighted meaningful genotype–phenotype interactionsas defined by three latent components showing a significant difference in the projection scores between patients and controls. Among the significant associations, the model highlighted EPHX1 and BCAS1 gene variant scores involved in neurodegenerative and myelination processes, hence relevant for AD. In particular, the first was associated with decreased subcortical volumes and the second with decreasedtemporal lobe thickness. Noteworthy, BCAS1 is particularly expressed in the dentate gyrus. Overall, the proposed approach allowed capturing genotype–phenotype interactions in a restricted study cohort that was confirmed by transcriptomic analysis, offering insights into the underlying mechanisms of neurodegeneration in AD in line with previous findings and suggesting new potential disease biomarkers.
中文翻译:

识别阿尔茨海默氏病脑萎缩和基因变异评分的联合特征
遗传数据和脑成像信息的联合建模可以确定阿尔茨海默病(AD)等神经退行性疾病此任务通常使用质量单变量方法来完成,这些方法依赖于一整套单核苷酸多态性 (SNP) 来评估它们与选定的图像衍生表型 (IDP) 的关联。然而,此类方法容易出现多重比较偏差,最重要的是,无法考虑潜在的跨特征交互,导致显着关联的检测不充分。克服这些限制同时减少性状数量的方法旨在在基因水平上传递遗传信息并捕获一组遗传变异的综合遗传效应,而不是单独观察每个 SNP。尽管它们可以揭示 AD 连续体中大脑调节的新的潜在遗传决定因素,但目前的文献仍然很大程度上未探讨它们与大脑 IDP 的关联。在这项工作中,我们依靠 AD 神经影像计划 (ADNI) 第 3 阶段数据集,探索了一种可解释的多元模型来分析灰质调节的遗传基础。T1 加权磁共振得出的皮质厚度和皮质下体积被认为可以描述成像表型。同时,遗传对应物由序列核关联测试(SKAT)过滤模型提取的基因变异得分来表示。此外,进行转录组分析结果强调了有意义的基因型-表型相互作用,由三个潜在成分定义,显示患者和对照之间的预测分数存在显着差异。在这些重要关联中,该模型强调了 EPHX1 和 BCAS1 基因变异评分参与神经退行性和髓鞘形成过程,因此与 AD 相关。特别是,第一个与皮质下体积减少有关,第二个与颞叶厚度减少有关。值得注意的是,BCAS1 在齿状回中特别表达。总体而言,所提出的方法允许在有限的研究队列,并通过转录组分析证实,根据以前的发现提供对 AD 神经退行性变的潜在机制的见解,并提出新的潜在疾病生物标志物。



























 京公网安备 11010802027423号
京公网安备 11010802027423号