Journal of Biomedical informatics ( IF 4.0 ) Pub Date : 2023-12-09 , DOI: 10.1016/j.jbi.2023.104568 Richard L Bradshaw 1 , Kensaku Kawamoto 1 , Jemar R Bather 2 , Melody S Goodman 2 , Wendy K Kohlmann 3 , Daniel Chavez-Yenter 4 , Molly Volkmar 5 , Rachel Monahan 6 , Kimberly A Kaphingst 4 , Guilherme Del Fiol 1
|
Objective
This study aimed to 1) investigate algorithm enhancements for identifying patients eligible for genetic testing of hereditary cancer syndromes using family history data from electronic health records (EHRs); and 2) assess their impact on relative differences across sex, race, ethnicity, and language preference.
Materials and Methods
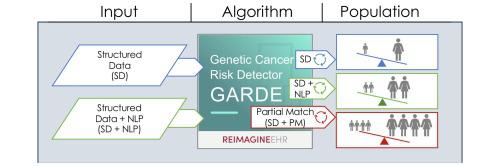
The study used EHR data from a tertiary academic medical center. A baseline rule-base algorithm, relying on structured family history data (structured data; SD), was enhanced using a natural language processing (NLP) component and a relaxed criteria algorithm (partial match [PM]). The identification rates and differences were analyzed considering sex, race, ethnicity, and language preference.
Results
Among 120,007 patients aged 25–60, detection rate differences were found across all groups using the SD (all P < 0.001). Both enhancements increased identification rates; NLP led to a 1.9 % increase and the relaxed criteria algorithm (PM) led to an 18.5 % increase (both P < 0.001). Combining SD with NLP and PM yielded a 20.4 % increase (P < 0.001). Similar increases were observed within subgroups. Relative differences persisted across most categories for the enhanced algorithms, with disproportionately higher identification of patients who are White, Female, non-Hispanic, and whose preferred language is English.
Conclusion
Algorithm enhancements increased identification rates for patients eligible for genetic testing of hereditary cancer syndromes, regardless of sex, race, ethnicity, and language preference. However, differences in identification rates persisted, emphasizing the need for additional strategies to reduce disparities such as addressing underlying biases in EHR family health information and selectively applying algorithm enhancements for disadvantaged populations. Systematic assessment of differences in algorithm performance across population subgroups should be incorporated into algorithm development processes.
中文翻译:

增强的基于家族史的算法增加了对符合遗传性癌症综合征基因检测标准的个体的识别,但本身并不能减少差异
客观的
本研究旨在 1) 研究算法增强,以使用电子健康记录 (EHR) 中的家族史数据来识别适合进行遗传性癌症综合征基因检测的患者; 2) 评估它们对性别、种族、民族和语言偏好之间相对差异的影响。
材料和方法
该研究使用了来自三级学术医疗中心的电子病历数据。使用自然语言处理 (NLP) 组件和宽松的标准算法(部分匹配 [PM])增强了依赖于结构化家族史数据(结构化数据;SD)的基线规则库算法。考虑性别、种族、民族和语言偏好来分析识别率和差异。
结果
在 120,007 名 25-60 岁患者中,使用 SD 发现所有组之间的检出率存在差异(所有 P < 0.001)。这两项增强功能都提高了识别率; NLP 导致 1.9% 的增长,宽松标准算法 (PM) 导致 18.5% 的增长(两者 P < 0.001)。将 SD 与 NLP 和 PM 相结合,可提高 20.4% (P < 0.001)。在亚组内也观察到类似的增加。增强算法的大多数类别都存在相对差异,对白人、女性、非西班牙裔且首选语言是英语的患者的识别率更高。
结论
算法的改进提高了符合遗传性癌症综合征基因检测资格的患者的识别率,无论性别、种族、民族和语言偏好如何。然而,识别率的差异仍然存在,强调需要采取额外的策略来减少差异,例如解决 EHR 家庭健康信息中的潜在偏差以及有选择地对弱势群体应用算法增强。对不同群体的算法性能差异的系统评估应该纳入算法开发过程中。











































 京公网安备 11010802027423号
京公网安备 11010802027423号