Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reverse transcription-free digital-quantitative-PCR for microRNA analysis
Analyst ( IF 3.6 ) Pub Date : 2023-05-31 , DOI: 10.1039/d3an00351e Hao T Mai 1 , Brice C Vanness 1 , Thomas H Linz 1
Analyst ( IF 3.6 ) Pub Date : 2023-05-31 , DOI: 10.1039/d3an00351e Hao T Mai 1 , Brice C Vanness 1 , Thomas H Linz 1
Affiliation
|
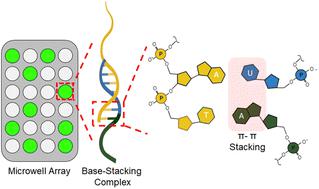
MicroRNAs (miRNAs) are non-coding RNA sequences that regulate many biological processes and have become central targets of biomedical research. However, their naturally low abundances in biological samples necessitates the development of sensitive analytical techniques to conduct routine miRNA measurements in research laboratories. Digital PCR has the potential to meet this need because of its single-molecule detection capabilities, but PCR analyses of miRNAs are slowed by the ligation and reverse transcription steps first required to prepare samples. This report describes the development of a method to rapidly quantify miRNA in digital microwell arrays using base-stacking digital-quantitative-PCR (BS-dqPCR). BS-dqPCR expedites miRNA measurements by eliminating the need for ligation and reverse transcription steps, which reduces the time and cost compared to conventional miRNA PCR analyses. Under standard PCR thermocycling conditions, digital signals from miRNA samples were lower than expected, while signals from blanks were high. Therefore, a novel asymmetric thermocycling program was developed that maximized on-target signal from miRNA while minimizing non-specific amplification. The analytical response of BS-dqPCR was then evaluated over a range of miRNA concentrations. The digital PCR dimension increased in signal with increasing miRNA copy numbers. When the digital signal saturated, the quantitative PCR dimension readily discerned miRNA copy number differences. Overall, BS-dqPCR provides rapid, high-sensitivity measurements of miRNA over a wide dynamic range, which demonstrates its utility for routine miRNA analyses.
中文翻译:

用于 microRNA 分析的无逆转录数字定量 PCR
MicroRNA (miRNA) 是调节许多生物过程的非编码 RNA 序列,已成为生物医学研究的中心目标。然而,它们在生物样品中的天然丰度较低,因此需要开发灵敏的分析技术来在研究实验室中进行常规 miRNA 测量。数字 PCR 因其单分子检测能力而有可能满足这一需求,但 miRNA 的 PCR 分析因制备样品首先需要的连接和逆转录步骤而减慢。本报告描述了一种使用碱基堆积数字定量 PCR (BS-dqPCR) 快速定量数字微孔阵列中 miRNA 的方法的开发。 BS-dqPCR 无需连接和逆转录步骤,从而加快了 miRNA 测量速度,与传统 miRNA PCR 分析相比,减少了时间和成本。在标准 PCR 热循环条件下,miRNA 样品的数字信号低于预期,而空白样品的信号较高。因此,开发了一种新颖的不对称热循环程序,可最大限度地提高 miRNA 的目标信号,同时最大限度地减少非特异性扩增。然后在一系列 miRNA 浓度范围内评估 BS-dqPCR 的分析响应。随着 miRNA 拷贝数的增加,数字 PCR 的信号强度也随之增加。当数字信号饱和时,定量 PCR 维度很容易辨别 miRNA 拷贝数差异。总体而言,BS-dqPCR 可在较宽的动态范围内快速、高灵敏度地测量 miRNA,这证明了其在常规 miRNA 分析中的实用性。
更新日期:2023-06-03
中文翻译:

用于 microRNA 分析的无逆转录数字定量 PCR
MicroRNA (miRNA) 是调节许多生物过程的非编码 RNA 序列,已成为生物医学研究的中心目标。然而,它们在生物样品中的天然丰度较低,因此需要开发灵敏的分析技术来在研究实验室中进行常规 miRNA 测量。数字 PCR 因其单分子检测能力而有可能满足这一需求,但 miRNA 的 PCR 分析因制备样品首先需要的连接和逆转录步骤而减慢。本报告描述了一种使用碱基堆积数字定量 PCR (BS-dqPCR) 快速定量数字微孔阵列中 miRNA 的方法的开发。 BS-dqPCR 无需连接和逆转录步骤,从而加快了 miRNA 测量速度,与传统 miRNA PCR 分析相比,减少了时间和成本。在标准 PCR 热循环条件下,miRNA 样品的数字信号低于预期,而空白样品的信号较高。因此,开发了一种新颖的不对称热循环程序,可最大限度地提高 miRNA 的目标信号,同时最大限度地减少非特异性扩增。然后在一系列 miRNA 浓度范围内评估 BS-dqPCR 的分析响应。随着 miRNA 拷贝数的增加,数字 PCR 的信号强度也随之增加。当数字信号饱和时,定量 PCR 维度很容易辨别 miRNA 拷贝数差异。总体而言,BS-dqPCR 可在较宽的动态范围内快速、高灵敏度地测量 miRNA,这证明了其在常规 miRNA 分析中的实用性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号