当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Shaping Nanobodies and Intrabodies against Proteoforms
Analytical Chemistry ( IF 7.4 ) Pub Date : 2023-05-26 , DOI: 10.1021/acs.analchem.3c00958 Bojana Leonard 1 , Vincent Danna 1 , Leo Gorham 1 , Michelle Davison 1 , William Chrisler 1 , Doo Nam Kim 1 , Vincent R Gerbasi 1
Analytical Chemistry ( IF 7.4 ) Pub Date : 2023-05-26 , DOI: 10.1021/acs.analchem.3c00958 Bojana Leonard 1 , Vincent Danna 1 , Leo Gorham 1 , Michelle Davison 1 , William Chrisler 1 , Doo Nam Kim 1 , Vincent R Gerbasi 1
Affiliation
|
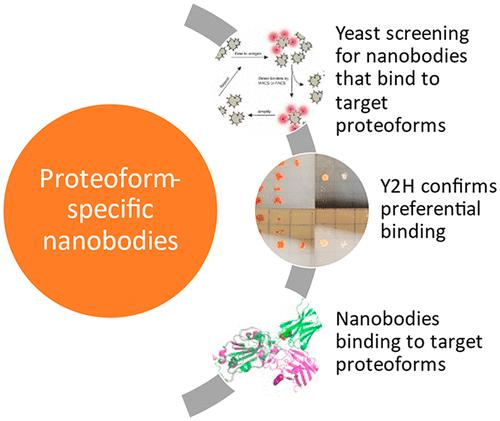
Proteoforms expand genomic diversity and direct developmental processes. While high-resolution mass spectrometry has accelerated characterization of proteoforms, molecular techniques working to bind and disrupt the function of specific proteoforms have lagged behind. In this study, we worked to develop intrabodies capable of binding specific proteoforms. We employed a synthetic camelid nanobody library expressed in yeast to identify nanobody binders of different SARS-CoV-2 receptor binding domain (RBD) proteoforms. Importantly, employment of the positive and negative selection mechanisms inherent to the synthetic system allowed for amplification of nanobody-expressing yeast that bind to the original (Wuhan strain RBD) but not the E484 K (Beta variant) mutation. Nanobodies raised against specific RBD proteoforms were validated by yeast-2-hybrid analysis and sequence comparisons. These results provide a framework for development of nanobodies and intrabodies that target proteoforms.
中文翻译:

针对蛋白质形态塑造纳米抗体和胞内抗体
Proteoforms 扩展基因组多样性和指导发展过程。虽然高分辨率质谱加速了蛋白质组的表征,但用于结合和破坏特定蛋白质组功能的分子技术却落后了。在这项研究中,我们致力于开发能够结合特定蛋白质组的胞内抗体。我们使用在酵母中表达的合成骆驼纳米抗体文库来鉴定不同 SARS-CoV-2 受体结合域 (RBD) 蛋白质组的纳米抗体结合剂。重要的是,利用合成系统固有的阳性和阴性选择机制,可以扩增与原始(武汉菌株 RBD)而非 E484 K(Beta 变体)突变结合的表达纳米抗体的酵母。针对特定 RBD 蛋白质形式产生的纳米抗体通过 yeast-2-hybrid 分析和序列比较进行了验证。这些结果为开发以蛋白质形式为目标的纳米抗体和胞内抗体提供了框架。
更新日期:2023-05-26
中文翻译:

针对蛋白质形态塑造纳米抗体和胞内抗体
Proteoforms 扩展基因组多样性和指导发展过程。虽然高分辨率质谱加速了蛋白质组的表征,但用于结合和破坏特定蛋白质组功能的分子技术却落后了。在这项研究中,我们致力于开发能够结合特定蛋白质组的胞内抗体。我们使用在酵母中表达的合成骆驼纳米抗体文库来鉴定不同 SARS-CoV-2 受体结合域 (RBD) 蛋白质组的纳米抗体结合剂。重要的是,利用合成系统固有的阳性和阴性选择机制,可以扩增与原始(武汉菌株 RBD)而非 E484 K(Beta 变体)突变结合的表达纳米抗体的酵母。针对特定 RBD 蛋白质形式产生的纳米抗体通过 yeast-2-hybrid 分析和序列比较进行了验证。这些结果为开发以蛋白质形式为目标的纳米抗体和胞内抗体提供了框架。



























 京公网安备 11010802027423号
京公网安备 11010802027423号