当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Dual UHPLC-HRMS Metabolomics and Lipidomics and Automated Data Processing Workflow for Comprehensive High-Throughput Gut Phenotyping
Analytical Chemistry ( IF 6.7 ) Pub Date : 2023-05-23 , DOI: 10.1021/acs.analchem.2c05371 P Vangeenderhuysen 1 , J Van Arnhem 1 , B Pomian 1 , M De Graeve 1 , L De Commer 2, 3 , G Falony 2, 3 , J Raes 2, 3 , A Zhernakova 4 , J Fu 4, 5 , L Y Hemeryck 1 , L Vanhaecke 1, 6
Analytical Chemistry ( IF 6.7 ) Pub Date : 2023-05-23 , DOI: 10.1021/acs.analchem.2c05371 P Vangeenderhuysen 1 , J Van Arnhem 1 , B Pomian 1 , M De Graeve 1 , L De Commer 2, 3 , G Falony 2, 3 , J Raes 2, 3 , A Zhernakova 4 , J Fu 4, 5 , L Y Hemeryck 1 , L Vanhaecke 1, 6
Affiliation
|
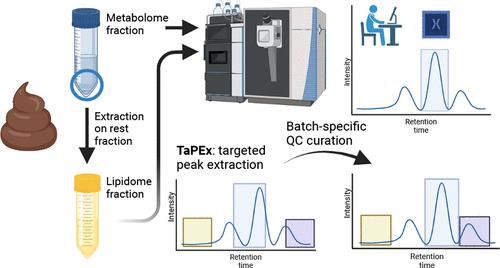
In recent years, feces has surfaced as the matrix of choice for investigating the gut microbiome-health axis because of its non-invasive sampling and the unique reflection it offers of an individual’s lifestyle. In cohort studies where the number of samples required is large, but availability is scarce, a clear need exists for high-throughput analyses. Such analyses should combine a wide physicochemical range of molecules with a minimal amount of sample and resources and downstream data processing workflows that are as automated and time efficient as possible. We present a dual fecal extraction and ultra high performance liquid chromatography-high resolution-quadrupole-orbitrap-mass spectrometry (UHPLC-HR-Q-Orbitrap-MS)-based workflow that enables widely targeted and untargeted metabolome and lipidome analysis. A total of 836 in-house standards were analyzed, of which 360 metabolites and 132 lipids were consequently detected in feces. Their targeted profiling was validated successfully with respect to repeatability (78% CV < 20%), reproducibility (82% CV < 20%), and linearity (81% R2 > 0.9), while also enabling holistic untargeted fingerprinting (15,319 features, CV < 30%). To automate targeted processing, we optimized an R-based targeted peak extraction (TaPEx) algorithm relying on a database comprising retention time and mass-to-charge ratio (360 metabolites and 132 lipids), with batch-specific quality control curation. The latter was benchmarked toward vendor-specific targeted and untargeted software and our isotopologue parameter optimization/XCMS-based untargeted pipeline in LifeLines Deep cohort samples (n = 97). TaPEx clearly outperformed the untargeted approaches (81.3 vs 56.7–66.0% compounds detected). Finally, our novel dual fecal metabolomics–lipidomics–TaPEx method was successfully applied to Flemish Gut Flora Project cohort (n = 292) samples, leading to a sample-to-result time reduction of 60%.
中文翻译:

双 UHPLC-HRMS 代谢组学和脂质组学以及用于综合高通量肠道表型分析的自动化数据处理工作流程
近年来,粪便已成为研究肠道微生物组-健康轴的首选基质,因为它是非侵入性采样,并且可以独特地反映个人的生活方式。在需要大量样本但可用性不足的队列研究中,显然需要高通量分析。此类分析应将广泛的物理化学分子范围与最少量的样品和资源以及尽可能自动化和省时的下游数据处理工作流程结合起来。我们提出了基于双重粪便提取和超高效液相色谱-高分辨率-四极杆-轨道阱-质谱 (UHPLC-HR-Q-Orbitrap-MS) 的工作流程,可实现广泛靶向和非靶向代谢组和脂质组分析。共分析了 836 种内部标准品,结果在粪便中检测到 360 种代谢物和 132 种脂质。他们的靶向分析在可重复性(78% CV < 20%)、再现性(82% CV < 20%)和线性(81%R 2 > 0.9),同时还支持整体非目标指纹识别(15,319 个特征,CV < 30%)。为了自动化靶向处理,我们优化了基于 R 的靶向峰提取 (TaPEx) 算法,该算法依赖于包含保留时间和质荷比(360 种代谢物和 132 种脂质)的数据库,以及批次特定的质量控制管理。后者针对供应商特定的目标和非目标软件以及我们在 LifeLines Deep 队列样本(n = 97)中的同位素异数体参数优化/基于 XCMS 的非目标管道进行了基准测试。TaPEx 明显优于非靶向方法(检测到 81.3 对 56.7–66.0% 的化合物)。最后,我们新的双重粪便代谢组学-脂质组学-TaPEx 方法成功应用于 Flemish Gut Flora Project 队列 ( n= 292) 个样本,从而使从样本到结果的时间减少了 60%。
更新日期:2023-05-23
中文翻译:

双 UHPLC-HRMS 代谢组学和脂质组学以及用于综合高通量肠道表型分析的自动化数据处理工作流程
近年来,粪便已成为研究肠道微生物组-健康轴的首选基质,因为它是非侵入性采样,并且可以独特地反映个人的生活方式。在需要大量样本但可用性不足的队列研究中,显然需要高通量分析。此类分析应将广泛的物理化学分子范围与最少量的样品和资源以及尽可能自动化和省时的下游数据处理工作流程结合起来。我们提出了基于双重粪便提取和超高效液相色谱-高分辨率-四极杆-轨道阱-质谱 (UHPLC-HR-Q-Orbitrap-MS) 的工作流程,可实现广泛靶向和非靶向代谢组和脂质组分析。共分析了 836 种内部标准品,结果在粪便中检测到 360 种代谢物和 132 种脂质。他们的靶向分析在可重复性(78% CV < 20%)、再现性(82% CV < 20%)和线性(81%R 2 > 0.9),同时还支持整体非目标指纹识别(15,319 个特征,CV < 30%)。为了自动化靶向处理,我们优化了基于 R 的靶向峰提取 (TaPEx) 算法,该算法依赖于包含保留时间和质荷比(360 种代谢物和 132 种脂质)的数据库,以及批次特定的质量控制管理。后者针对供应商特定的目标和非目标软件以及我们在 LifeLines Deep 队列样本(n = 97)中的同位素异数体参数优化/基于 XCMS 的非目标管道进行了基准测试。TaPEx 明显优于非靶向方法(检测到 81.3 对 56.7–66.0% 的化合物)。最后,我们新的双重粪便代谢组学-脂质组学-TaPEx 方法成功应用于 Flemish Gut Flora Project 队列 ( n= 292) 个样本,从而使从样本到结果的时间减少了 60%。











































 京公网安备 11010802027423号
京公网安备 11010802027423号