当前位置:
X-MOL 学术
›
J. Phys. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deep Boosted Molecular Dynamics: Accelerating Molecular Simulations with Gaussian Boost Potentials Generated Using Probabilistic Bayesian Deep Neural Network
The Journal of Physical Chemistry Letters ( IF 4.8 ) Pub Date : 2023-05-23 , DOI: 10.1021/acs.jpclett.3c00926 Hung N Do 1 , Yinglong Miao 1
The Journal of Physical Chemistry Letters ( IF 4.8 ) Pub Date : 2023-05-23 , DOI: 10.1021/acs.jpclett.3c00926 Hung N Do 1 , Yinglong Miao 1
Affiliation
|
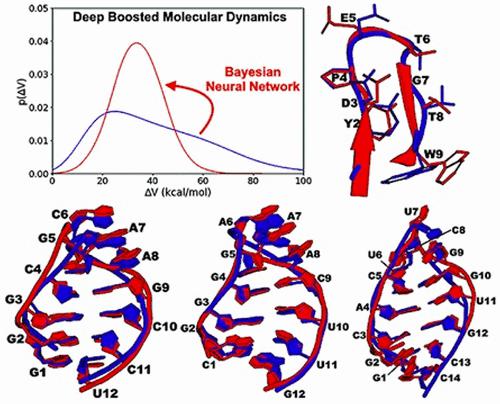
We have developed a new deep boosted molecular dynamics (DBMD) method. Probabilistic Bayesian neural network models were implemented to construct boost potentials that exhibit Gaussian distribution with minimized anharmonicity, thereby allowing for accurate energetic reweighting and enhanced sampling of molecular simulations. DBMD was demonstrated on model systems of alanine dipeptide and the fast-folding protein and RNA structures. For alanine dipeptide, 30 ns DBMD simulations captured up to 83–125 times more backbone dihedral transitions than 1 μs conventional molecular dynamics (cMD) simulations and were able to accurately reproduce the original free energy profiles. Moreover, DBMD sampled multiple folding and unfolding events within 300 ns simulations of the chignolin model protein and identified low-energy conformational states comparable to previous simulation findings. Finally, DBMD captured a general folding pathway of three hairpin RNAs with the GCAA, GAAA, and UUCG tetraloops. Based on a deep learning neural network, DBMD provides a powerful and generally applicable approach to boosting biomolecular simulations. DBMD is available with open source in OpenMM at https://github.com/MiaoLab20/DBMD/.
中文翻译:

深度增强分子动力学:利用概率贝叶斯深度神经网络生成的高斯增强势加速分子模拟
我们开发了一种新的深度增强分子动力学(DBMD)方法。采用概率贝叶斯神经网络模型来构建表现出具有最小化非和谐性的高斯分布的增强势,从而允许精确的能量重新加权和增强分子模拟的采样。DBMD 在丙氨酸二肽以及快速折叠蛋白和 RNA 结构的模型系统上得到了证明。对于丙氨酸二肽,30 ns DBMD 模拟捕获的主链二面角跃迁比 1 μs 传统分子动力学 (cMD) 模拟多 83-125 倍,并且能够准确再现原始自由能分布。此外,DBMD 在 chignolin 模型蛋白的 300 ns 模拟内对多个折叠和展开事件进行了采样,并确定了与之前的模拟结果相当的低能构象状态。最后,DBMD 捕获了具有 GCAA、GAAA 和 UUCG 四环的三种发夹 RNA 的一般折叠途径。DBMD 基于深度学习神经网络,提供了一种强大且普遍适用的方法来促进生物分子模拟。DBMD 在 OpenMM 中以开源形式提供,网址为 https://github.com/MiaoLab20/DBMD/。
更新日期:2023-05-23
中文翻译:

深度增强分子动力学:利用概率贝叶斯深度神经网络生成的高斯增强势加速分子模拟
我们开发了一种新的深度增强分子动力学(DBMD)方法。采用概率贝叶斯神经网络模型来构建表现出具有最小化非和谐性的高斯分布的增强势,从而允许精确的能量重新加权和增强分子模拟的采样。DBMD 在丙氨酸二肽以及快速折叠蛋白和 RNA 结构的模型系统上得到了证明。对于丙氨酸二肽,30 ns DBMD 模拟捕获的主链二面角跃迁比 1 μs 传统分子动力学 (cMD) 模拟多 83-125 倍,并且能够准确再现原始自由能分布。此外,DBMD 在 chignolin 模型蛋白的 300 ns 模拟内对多个折叠和展开事件进行了采样,并确定了与之前的模拟结果相当的低能构象状态。最后,DBMD 捕获了具有 GCAA、GAAA 和 UUCG 四环的三种发夹 RNA 的一般折叠途径。DBMD 基于深度学习神经网络,提供了一种强大且普遍适用的方法来促进生物分子模拟。DBMD 在 OpenMM 中以开源形式提供,网址为 https://github.com/MiaoLab20/DBMD/。











































 京公网安备 11010802027423号
京公网安备 11010802027423号