Phytomedicine ( IF 6.7 ) Pub Date : 2023-05-13 , DOI: 10.1016/j.phymed.2023.154882 Xiaoxia Ding 1 , Jieting Chen 1 , Chunyan Dai 1 , Peiqi Shi 1 , Hengyu Pan 1 , Yanqi Lin 1 , Yikang Chen 1 , Lu Gong 1 , Linming Chen 2 , Wenguang Wu 1 , Xiaohui Qiu 1 , Jiang Xu 3 , Zhihai Huang 1 , Baosheng Liao 1
|
Background
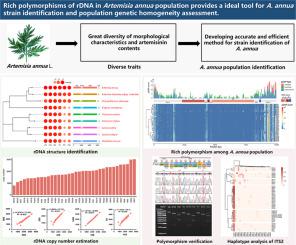
Artemisia annua, a well-known traditional Chinese medicine, is the main source for production of artemisinin, an anti-malaria drug. A. annua is distributed globally, with great diversity of morphological characteristics and artemisinin contents. Diverse traits among A. annua populations impeded the stable production of artemisinin, which needs an efficient tool to identify strains and assess population genetic homogeneity.
Purpose
In this study, ribosomal DNA (rDNA), were characterized for A. annua for strains identification and population genetic homogeneity assessment.
Methods
The ribosomal RNA (rRNA) genes were identified using cmscan and assembled using rDNA unit of LQ-9 as a reference. rDNA among Asteraceae species were compared performing with 45S rDNA. The rDNA copy number was calculated based on sequencing depth. The polymorphisms of rDNA sequences were identified with bam-readcount, and confirmed by Sanger sequencing and restriction enzyme experiment. The ITS2 amplicon sequencing was used to verify the stability of ITS2 haplotype analysis.
Results
Different from other Asteraceae species, 45S and 5S linked-type rDNA was only found in Artemisia genus. Rich polymorphisms of copy number and sequence of rDNA were identified in A. annua population. The haplotype composition of internal transcribed spacer 2 (ITS2) region which had moderate sequence polymorphism and relative short size was significantly different among A. annua strains. A population discrimination method was developed based on ITS2 haplotype analysis with high-throughput sequencing.
Conclusion
This study provides comprehensive characteristics of rDNA and suggests that ITS2 haplotype analysis is ideal tool for A. annua strain identification and population genetic homogeneity assessment.
中文翻译:

开发基于rDNA多态性的中药种群鉴定工具:青蒿。
背景
青蒿是一种著名的中药,是生产抗疟疾药物青蒿素的主要来源。青蒿全球分布,形态特征和青蒿素含量差异很大。青蒿种群的不同性状阻碍了青蒿素的稳定生产,这需要一种有效的工具来鉴定菌株和评估种群遗传同质性。
目的
在这项研究中,对青蒿的核糖体 DNA (rDNA) 进行了鉴定,以用于菌株鉴定和种群遗传同质性评估。
方法
使用 cmscan 识别核糖体 RNA (rRNA) 基因,并使用 LQ-9 的 rDNA 单元作为参考进行组装。将菊科物种中的 rDNA 与 45S rDNA 进行比较。根据测序深度计算 rDNA 拷贝数。rDNA序列的多态性通过bam-readcount进行鉴定,并通过Sanger测序和酶切实验加以证实。ITS2扩增子测序用于验证ITS2单倍型分析的稳定性。
结果
与其他菊科植物不同,45S和5S连锁型rDNA仅在蒿属植物中发现。在青蒿群体中鉴定出丰富的rDNA拷贝数和序列多态性。具有中等序列多态性和相对短尺寸的内部转录间隔区2(ITS2)区域的单倍型组成在青蒿菌株之间存在显着差异。基于 ITS2 单倍型分析和高通量测序开发了一种群体区分方法。
结论
本研究提供了 rDNA 的综合特征,表明 ITS2 单倍型分析是青蒿菌株鉴定和种群遗传同质性评估的理想工具。











































 京公网安备 11010802027423号
京公网安备 11010802027423号