Structure ( IF 4.4 ) Pub Date : 2023-05-10 , DOI: 10.1016/j.str.2023.04.007 Philipp Junk 1 , Christina Kiel 2
|
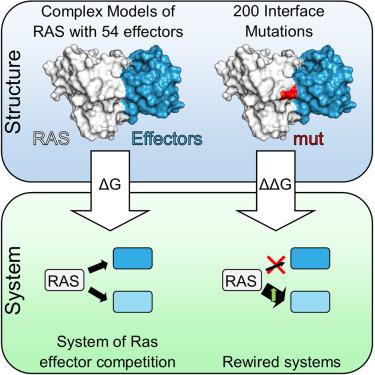
Ras is a central cellular hub protein controlling multiple cell fates. How Ras interacts with a variety of potential effector proteins is relatively unexplored, with only some key effectors characterized in great detail. Here, we have used homology modeling based on X-ray and AlphaFold2 templates to build structural models for 54 Ras-effector complexes. These models were used to estimate binding affinities using a supervised learning regressor. Furthermore, we systematically introduced Ras “branch-pruning” (or branchegetic) mutations to identify 200 interface mutations that affect the binding energy with at least one of the model structures. The impacts of these branchegetic mutants were integrated into a mathematical model to assess the potential for rewiring interactions at the Ras hub on a systems level. These findings have provided a quantitative understanding of Ras-effector interfaces and their impact on systems properties of a key cellular hub.
中文翻译:

基于结构的 Ras 效应子结合亲和力预测和“分支”界面突变的设计
Ras 是控制多种细胞命运的细胞中心蛋白。 Ras 如何与各种潜在效应蛋白相互作用尚未被探索,仅对一些关键效应蛋白进行了详细描述。在这里,我们使用基于 X 射线和 AlphaFold2 模板的同源建模来构建 54 个 Ras 效应器复合物的结构模型。这些模型用于使用监督学习回归器来估计结合亲和力。此外,我们系统地引入了 Ras“分支修剪”(或分支)突变,以识别 200 个影响与至少一种模型结构的结合能的界面突变。这些分支突变体的影响被整合到一个数学模型中,以评估 Ras 中心在系统水平上重新布线相互作用的潜力。这些发现提供了对 Ras 效应器接口及其对关键细胞中枢系统特性的影响的定量理解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号