Structure ( IF 4.4 ) Pub Date : 2023-03-23 , DOI: 10.1016/j.str.2023.03.001 Eva M Huber 1 , Lukas Kreling 2 , Antje K Heinrich 2 , Maximilian Dünnebacke 1 , Alexander Pöthig 3 , Helge B Bode 4 , Michael Groll 1
|
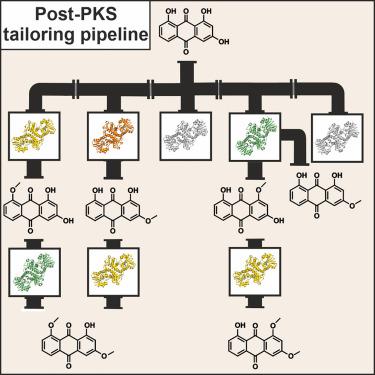
Modification of the polyketide anthraquinone AQ-256 in the entomopathogenic Photorhabdus luminescens involves several O-methylations, but the biosynthetic gene cluster antA-I lacks corresponding tailoring enzymes. We here describe the identification of five putative, highly homologous O-methyltransferases encoded in the genome of P. luminescens. Activity assays in vitro and deletion experiments in vivo revealed that three of them account for anthraquinone tailoring by producing three monomethylated and two dimethylated species of AQ-256. X-ray structures of all five enzymes indicate high structural and mechanistic similarity. As confirmed by structure-based mutagenesis, a conserved histidine at the active site likely functions as a general base for substrate deprotonation and subsequent methyl transfer in all enzymes. Eight complex structures with AQ-256 as well as mono- and dimethylated derivatives confirm the substrate specificity patterns found in vitro and visualize how single amino acid differences in the active-site pockets impact substrate orientation and govern site-specific methylation.
中文翻译:

一组密切相关的甲基转移酶,用于蒽醌色素的位点特异性剪裁
昆虫病原 发光杆菌中的聚酮化合物蒽醌 AQ-256 的修饰涉及多个O甲基化,但生物合成基因簇antA-I缺乏相应的剪裁酶。我们在这里描述了在P. luminescens的基因组中编码的五种推定的,高度同源的O -甲基转移酶的鉴定。体外活性测定和体内缺失实验揭示了其中三个通过产生三种单甲基化和两种二甲基化的 AQ-256 来解释蒽醌剪裁。所有五种酶的 X 射线结构表明结构和机制具有高度相似性。正如基于结构的诱变所证实的那样,活性位点处的保守组氨酸可能作为底物去质子化和所有酶中随后的甲基转移的通用碱基。具有 AQ-256 的八种复杂结构以及单甲基化和二甲基化衍生物证实了体外发现的底物特异性模式,并可视化了活性位点口袋中的单个氨基酸差异如何影响底物方向和控制位点特异性甲基化。











































 京公网安备 11010802027423号
京公网安备 11010802027423号