Cell ( IF 45.5 ) Pub Date : 2022-12-22 , DOI: 10.1016/j.cell.2022.11.028 Mohamed Ameen 1 , Laksshman Sundaram 2 , Mengcheng Shen 3 , Abhimanyu Banerjee 4 , Soumya Kundu 5 , Surag Nair 5 , Anna Shcherbina 6 , Mingxia Gu 7 , Kitchener D Wilson 3 , Avyay Varadarajan 8 , Nirmal Vadgama 9 , Akshay Balsubramani 10 , Joseph C Wu 3 , Jesse M Engreitz 10 , Kyle Farh 11 , Ioannis Karakikes 12 , Kevin C Wang 13 , Thomas Quertermous 14 , William J Greenleaf 15 , Anshul Kundaje 16
|
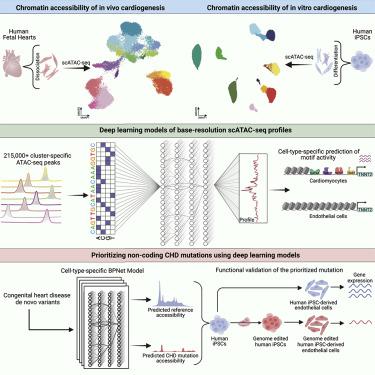
To define the multi-cellular epigenomic and transcriptional landscape of cardiac cellular development, we generated single-cell chromatin accessibility maps of human fetal heart tissues. We identified eight major differentiation trajectories involving primary cardiac cell types, each associated with dynamic transcription factor (TF) activity signatures. We contrasted regulatory landscapes of iPSC-derived cardiac cell types and their in vivo counterparts, which enabled optimization of in vitro differentiation of epicardial cells. Further, we interpreted sequence based deep learning models of cell-type-resolved chromatin accessibility profiles to decipher underlying TF motif lexicons. De novo mutations predicted to affect chromatin accessibility in arterial endothelium were enriched in congenital heart disease (CHD) cases vs. controls. In vitro studies in iPSCs validated the functional impact of identified variation on the predicted developmental cell types. This work thus defines the cell-type-resolved cis-regulatory sequence determinants of heart development and identifies disruption of cell type-specific regulatory elements in CHD.
中文翻译:

心脏发生的综合单细胞分析确定先天性心脏病的发育轨迹和非编码突变
为了定义心脏细胞发育的多细胞表观基因组和转录景观,我们生成了人类胎儿心脏组织的单细胞染色质可及性图。我们确定了涉及原代心肌细胞类型的八种主要分化轨迹,每种轨迹都与动态转录因子(TF)活性特征相关。我们对比了 iPSC 衍生的心脏细胞类型和体内对应细胞的调控景观,从而优化了心外膜细胞的体外分化。此外,我们解释了基于序列的细胞类型解析染色质可及性特征的深度学习模型,以破译潜在的 TF 基序词典。与对照组相比,先天性心脏病 (CHD) 病例中预测会影响动脉内皮染色质可及性的新生突变较多。 iPSC 的体外研究验证了已识别变异对预测发育细胞类型的功能影响。因此,这项工作定义了心脏发育的细胞类型解析的顺式调节序列决定因素,并确定了 CHD 中细胞类型特异性调节元件的破坏。











































 京公网安备 11010802027423号
京公网安备 11010802027423号