Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2022-12-02 , DOI: 10.1016/j.csbj.2022.11.053 Ina Bang 1 , Linh Khanh Nong 1 , Joon Young Park 1 , Hoa Thi Le 1 , Sang- Mok Lee 1 , Donghyuk Kim 1, 2
|
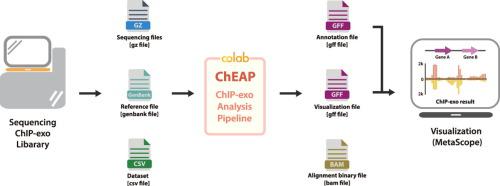
Genome-scale studies of the bacterial regulatory network have been leveraged by declining sequencing cost and advances in ChIP (chromatin immunoprecipitation) methods. Of which, ChIP-exo has proven competent with its near-single base-pair resolution. While several algorithms and programs have been developed for different analytical steps in ChIP-exo data processing, there is a lack of effort in incorporating them into a convenient bioinformatics pipeline that is intuitive and publicly available. In this paper, we developed ChIP-exo Analysis Pipeline (ChEAP) that executes the one-step process, starting from trimming and aligning raw sequencing reads to visualization of ChIP-exo results. The pipeline was implemented on the interactive web-based Python development environment – Jupyter Notebook, which is compatible with the Google Colab cloud platform to facilitate the sharing of codes and collaboration among researchers. Additionally, users could exploit the free GPU and CPU resources allocated by Colab to carry out computing tasks regardless of the performance of their local machines. The utility of ChEAP was demonstrated with the ChIP-exo datasets of RpoN sigma factor in E. coli K-12 MG1655. To analyze two raw data files, ChEAP runtime was 2 min and 25 s. Subsequent analyses identified 113 RpoN binding sites showing a conserved RpoN binding pattern in the motif search. ChEAP application in ChIP-exo data analysis is extensive and flexible for the parallel processing of data from various organisms.
中文翻译:

ChEAP:ChIP-exo 分析流程和大肠杆菌 RpoN 蛋白-DNA 相互作用的研究
通过降低测序成本和 ChIP(染色质免疫沉淀)方法的进步,细菌调控网络的基因组规模研究得到了利用。其中,ChIP-exo 已被证明具有近乎单碱基对分辨率的能力。虽然已经针对 ChIP-exo 数据处理中的不同分析步骤开发了多种算法和程序,但仍缺乏将它们整合到直观且公开可用的便捷生物信息学管道中的努力。在本文中,我们开发了 ChIP-exo 分析管道 (ChEAP),该管道执行一步过程,从修剪和对齐原始测序读数到 ChIP-exo 结果的可视化。该管道是在基于 Web 的交互式 Python 开发环境——Jupyter Notebook 上实现的,兼容谷歌Colab云平台,方便研究人员之间的代码共享和协作。此外,无论本地机器的性能如何,用户都可以利用 Colab 分配的免费 GPU 和 CPU 资源来执行计算任务。ChEAP 的实用性已通过 RpoN 西格玛因子的 ChIP-exo 数据集在大肠杆菌K-12 MG1655。为了分析两个原始数据文件,ChEAP 运行时间为 2 分钟 25 秒。随后的分析确定了 113 个 RpoN 结合位点,在基序搜索中显示出保守的 RpoN 结合模式。ChEAP 在 ChIP-exo 数据分析中的应用广泛且灵活,可以并行处理来自不同生物体的数据。









































 京公网安备 11010802027423号
京公网安备 11010802027423号