Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2022-12-01 , DOI: 10.1016/j.csbj.2022.11.057 Sumin Lee 1 , Seeun Kim 2 , Gyu Rie Lee 3 , Sohee Kwon 2 , Hyeonuk Woo 2 , Chaok Seok 2 , Hahnbeom Park 4
|
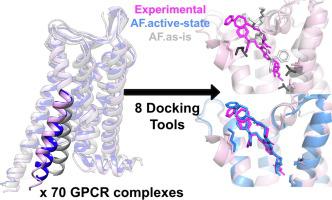
While deep learning (DL) has brought a revolution in the protein structure prediction field, still an important question remains how the revolution can be transferred to advances in structure-based drug discovery. Because the lessons from the recent GPCR dock challenge were inconclusive primarily due to the size of the dataset, in this work we further elaborated on 70 diverse GPCR complexes bound to either small molecules or peptides to investigate the best-practice modeling and docking strategies for GPCR drug discovery. From our quantitative analysis, it is shown that substantial improvements in docking and virtual screening have been possible by the advance in DL-based protein structure predictions with respect to the expected results from the combination of best pre-DL tools. The success rate of docking on DL-based model structures approaches that of cross-docking on experimental structures, showing over 30% improvement from the best pre-DL protocols. This amount of performance could be achieved only when two modeling points were considered properly: 1) correct functional-state modeling of receptors and 2) receptor-flexible docking. Best-practice modeling strategies and the model confidence estimation metric suggested in this work may serve as a guideline for future computer-aided GPCR drug discovery scenarios.
中文翻译:

评估基于深度学习的蛋白质结构预测时代的 GPCR 建模和对接策略
虽然深度学习(DL)给蛋白质结构预测领域带来了一场革命,但如何将这场革命转化为基于结构的药物发现的进步仍然是一个重要的问题。由于最近的 GPCR 对接挑战的经验教训主要由于数据集的大小而不确定,因此在这项工作中,我们进一步详细阐述了与小分子或肽结合的 70 种不同的 GPCR 复合物,以研究 GPCR 的最佳实践建模和对接策略药物发现。我们的定量分析表明,基于深度学习的蛋白质结构预测的进步,相对于最佳预深度学习工具组合的预期结果,可以在对接和虚拟筛选方面取得实质性改进。基于 DL 的模型结构对接的成功率接近实验结构交叉对接的成功率,与最佳的 DL 前协议相比提高了 30% 以上。只有正确考虑两个建模点才能实现这种性能:1)正确的受体功能状态建模和2)受体灵活对接。这项工作中建议的最佳实践建模策略和模型置信度估计指标可以作为未来计算机辅助 GPCR 药物发现场景的指南。











































 京公网安备 11010802027423号
京公网安备 11010802027423号