当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Exploration of Chemical Space Guided by PixelCNN for Fragment-Based De Novo Drug Discovery
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-12-01 , DOI: 10.1021/acs.jcim.2c01345 Satoshi Noguchi 1 , Junya Inoue 2, 3, 4
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-12-01 , DOI: 10.1021/acs.jcim.2c01345 Satoshi Noguchi 1 , Junya Inoue 2, 3, 4
Affiliation
|
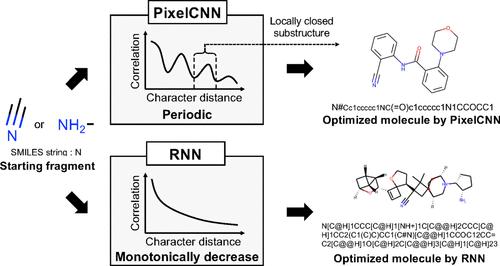
We report a novel framework for achieving fragment-based molecular design using pixel convolutional neural network (PixelCNN) combined with the simplified molecular input line entry system (SMILES) as molecular representation. While a widely used recurrent neural network (RNN) assumes monotonically decaying correlations in strings, PixelCNN captures a periodicity among characters of SMILES. Thus, PixelCNN provides us with a novel solution for the analysis of chemical space by extracting the periodicity of molecular structures that will be buried in SMILES. Moreover, this characteristic enables us to generate molecules by combining several simple building blocks, such as a benzene ring and side-chain structures, which contributes to the effective exploration of chemical space by step-by-step searching for molecules from a target fragment. In conclusion, PixelCNN could be a powerful approach focusing on the periodicity of molecules to explore chemical space for the fragment-based molecular design.
中文翻译:

PixelCNN 引导的化学空间探索,用于基于片段的从头药物发现
我们报告了一种使用像素卷积神经网络 (PixelCNN) 结合简化分子输入行输入系统 (SMILES) 作为分子表示来实现基于片段的分子设计的新框架。虽然广泛使用的递归神经网络 (RNN) 假设字符串中存在单调衰减的相关性,但 PixelCNN 捕获了 SMILES 字符之间的周期性。因此,PixelCNN 通过提取将被掩埋在 SMILES 中的分子结构的周期性,为我们提供了一种新的化学空间分析解决方案。此外,这一特性使我们能够通过组合几个简单的构建单元(例如苯环和侧链结构)来生成分子,这有助于通过从目标片段逐步搜索分子来有效探索化学空间。
更新日期:2022-12-01
中文翻译:

PixelCNN 引导的化学空间探索,用于基于片段的从头药物发现
我们报告了一种使用像素卷积神经网络 (PixelCNN) 结合简化分子输入行输入系统 (SMILES) 作为分子表示来实现基于片段的分子设计的新框架。虽然广泛使用的递归神经网络 (RNN) 假设字符串中存在单调衰减的相关性,但 PixelCNN 捕获了 SMILES 字符之间的周期性。因此,PixelCNN 通过提取将被掩埋在 SMILES 中的分子结构的周期性,为我们提供了一种新的化学空间分析解决方案。此外,这一特性使我们能够通过组合几个简单的构建单元(例如苯环和侧链结构)来生成分子,这有助于通过从目标片段逐步搜索分子来有效探索化学空间。










































 京公网安备 11010802027423号
京公网安备 11010802027423号