当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
PPI-Miner: A Structure and Sequence Motif Co-Driven Protein–Protein Interaction Mining and Modeling Computational Method
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-30 , DOI: 10.1021/acs.jcim.2c01033 Lin Wang , Feng-Lei Li , Xin-Yue Ma , Yong Cang , Fang Bai 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-30 , DOI: 10.1021/acs.jcim.2c01033 Lin Wang , Feng-Lei Li , Xin-Yue Ma , Yong Cang , Fang Bai 1
Affiliation
|
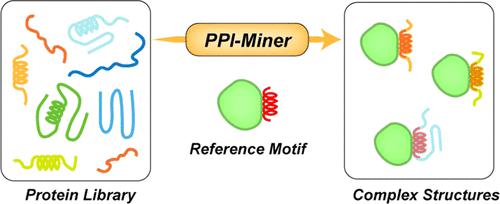
Protein–protein interactions (PPIs) play important roles in biological processes of life, and predicting PPIs becomes a critical scientific issue of concern. Most PPIs occur through small domains or motifs (fragments), which are challenging and laborious to map by standard biochemical approaches because they generally require the cloning of several truncation mutants. Here, we present a computational method, named as PPI-Miner, to fish potential protein interacting partners utilizing protein motifs as queries. In brief, this work first developed a motif-matching algorithm designed to identify the proteins that contain sequential or structural similar motifs with the given query motif. Being aligned to the query motif, the binding mode of the discovered motif and its receptor protein will be initially determined to be used to build PPI complexes accordingly. Eventually, a PPI complex structure could be built and optimized with a designed automatic protocol. Besides discovering PPIs, PPI-Miner can also be applied to other areas, i.e., the rational design of molecular glues and protein vaccines. In this work, PPI-Miner was employed to mine the potential cereblon (CRBN) substrates from human proteome. As a result, 1,739 candidates were predicted, and 16 of them have been experimentally validated in previous studies. The source code of PPI-Miner can be obtained from the GitHub repository (https://github.com/Wang-Lin-boop/PPI-Miner), the webserver is freely available for users (https://bailab.siais.shanghaitech.edu.cn/services/ppi-miner), and the database of predicted CRBN substrates is accessible at https://bailab.siais.shanghaitech.edu.cn/services/crbn-subslib.
中文翻译:

PPI-Miner:一种结构和序列基序共同驱动的蛋白质-蛋白质相互作用挖掘和建模计算方法
蛋白质-蛋白质相互作用 (PPI) 在生命的生物过程中起着重要作用,预测 PPI 成为一个关键的科学问题。大多数 PPI 通过小结构域或基序(片段)发生,通过标准生化方法进行映射具有挑战性和费力,因为它们通常需要克隆多个截断突变体。在这里,我们提出了一种名为 PPI-Miner 的计算方法,以利用蛋白质基序作为查询来捕获潜在的蛋白质相互作用伙伴。简而言之,这项工作首先开发了一种基序匹配算法,旨在识别包含具有给定查询基序的顺序或结构相似基序的蛋白质。与查询主题对齐,初步确定所发现的基序与其受体蛋白的结合方式,用于构建相应的 PPI 复合物。最终,可以使用设计的自动协议构建和优化 PPI 复杂结构。除了发现PPIs,PPI-Miner还可以应用于其他领域,即分子胶和蛋白质疫苗的合理设计。在这项工作中,PPI-Miner 被用来从人类蛋白质组中挖掘潜在的 cereblon (CRBN) 底物。结果,预测了 1,739 名候选人,其中 16 名已在先前的研究中通过实验验证。PPI-Miner 的源代码可以从 GitHub 存储库 (https://github.com/Wang-Lin-boop/PPI-Miner) 获得,网络服务器免费提供给用户 (https://bailab.siais. shanghaitech.edu.cn/services/ppi-miner),
更新日期:2022-11-30
中文翻译:

PPI-Miner:一种结构和序列基序共同驱动的蛋白质-蛋白质相互作用挖掘和建模计算方法
蛋白质-蛋白质相互作用 (PPI) 在生命的生物过程中起着重要作用,预测 PPI 成为一个关键的科学问题。大多数 PPI 通过小结构域或基序(片段)发生,通过标准生化方法进行映射具有挑战性和费力,因为它们通常需要克隆多个截断突变体。在这里,我们提出了一种名为 PPI-Miner 的计算方法,以利用蛋白质基序作为查询来捕获潜在的蛋白质相互作用伙伴。简而言之,这项工作首先开发了一种基序匹配算法,旨在识别包含具有给定查询基序的顺序或结构相似基序的蛋白质。与查询主题对齐,初步确定所发现的基序与其受体蛋白的结合方式,用于构建相应的 PPI 复合物。最终,可以使用设计的自动协议构建和优化 PPI 复杂结构。除了发现PPIs,PPI-Miner还可以应用于其他领域,即分子胶和蛋白质疫苗的合理设计。在这项工作中,PPI-Miner 被用来从人类蛋白质组中挖掘潜在的 cereblon (CRBN) 底物。结果,预测了 1,739 名候选人,其中 16 名已在先前的研究中通过实验验证。PPI-Miner 的源代码可以从 GitHub 存储库 (https://github.com/Wang-Lin-boop/PPI-Miner) 获得,网络服务器免费提供给用户 (https://bailab.siais. shanghaitech.edu.cn/services/ppi-miner),











































 京公网安备 11010802027423号
京公网安备 11010802027423号