当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
KID: A Kinase-Focused Interaction Database and Its Application in the Construction of Kinase-Focused Molecule Databases
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-29 , DOI: 10.1021/acs.jcim.2c00908 Xiaowen Dai 1 , Yuan Xu 1 , Haodi Qiu 1 , Xu Qian 1 , Mingde Lin 1 , Lin Luo 1 , Yang Zhao 1 , Dingfang Huang 1 , Yanmin Zhang 1 , Yadong Chen 1 , Haichun Liu 1 , Yulei Jiang 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-29 , DOI: 10.1021/acs.jcim.2c00908 Xiaowen Dai 1 , Yuan Xu 1 , Haodi Qiu 1 , Xu Qian 1 , Mingde Lin 1 , Lin Luo 1 , Yang Zhao 1 , Dingfang Huang 1 , Yanmin Zhang 1 , Yadong Chen 1 , Haichun Liu 1 , Yulei Jiang 1
Affiliation
|
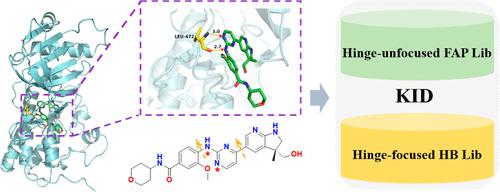
Protein kinases are important drug targets for the treatment of several diseases. The interaction between kinases and ligands is vital in the process of small-molecule kinase inhibitor (SMKI) design. In this study, we propose a method to extract fragments and amino acid residues from crystal structures for kinase–ligand interactions. In addition, core fragments that interact with the important hinge region of kinases were extracted along with their decorations. Based on the superimposed structural data of kinases from the kinase–ligand interaction fingerprint and structure database, we obtained two libraries, namely, a hinge-unfocused fragment–amino acid pair library (FAP Lib) that contains 6672 pairs of fragments and corresponding amino-acids, and a hinge-focused hinge binder library (HB Lib) of 3560 pairs of hinge-binding scaffolds with their corresponding decorations. These two libraries constitute a kinase-focused interaction database (KID). In depth analysis was conducted on KID to explore important characteristics of fragments in the design of SMKIs. With KID, we built two kinase-focused molecule databases, one called Recomb_DB, which contains 1,72,346 molecules generated through fragment recombination based on the FAP Lib, and another called RsdHB_DB, which contains 93,030 molecules generated based on our HB Lib using molecular generation methods. Compared with five databases both commercial and non-commercial, these two databases both ranked top 3 in scaffold diversity, top 4 in molecule fingerprint diversity, and are more focused on the chemical space of kinase inhibitors. Hence, KID presents a useful addition to existing databases for the exploration of novel SMKIs.
中文翻译:

KID:一个以激酶为中心的相互作用数据库及其在以激酶为中心的分子数据库构建中的应用
蛋白激酶是治疗多种疾病的重要药物靶标。激酶和配体之间的相互作用在小分子激酶抑制剂 (SMKI) 设计过程中至关重要。在这项研究中,我们提出了一种从晶体结构中提取片段和氨基酸残基以进行激酶-配体相互作用的方法。此外,还提取了与激酶重要铰链区相互作用的核心片段及其装饰物。基于来自激酶-配体相互作用指纹和结构数据库的激酶的叠加结构数据,我们获得了两个文库,即包含 6672 对片段和相应氨基-的铰链非聚焦片段-氨基酸对文库 (FAP Lib)。酸,和一个以铰链为中心的铰链活页夹库(HB Lib),包含 3560 对铰链绑定支架及其相应的装饰。这两个库构成了一个以激酶为中心的相互作用数据库 (KID)。对 KID 进行了深入分析,以探索片段在 SMKI 设计中的重要特征。借助 KID,我们构建了两个以激酶为中心的分子数据库,一个名为 Recomb_DB,其中包含基于 FAP Lib 通过片段重组生成的 1,72,346 个分子,另一个名为 RsdHB_DB,其中包含基于我们的 HB Lib 使用分子生成生成的 93,030 个分子方法。与商业和非商业的五个数据库相比,这两个数据库在支架多样性方面均排名前三,在分子指纹多样性方面均排名前四,并且更侧重于激酶抑制剂的化学空间。
更新日期:2022-11-29
中文翻译:

KID:一个以激酶为中心的相互作用数据库及其在以激酶为中心的分子数据库构建中的应用
蛋白激酶是治疗多种疾病的重要药物靶标。激酶和配体之间的相互作用在小分子激酶抑制剂 (SMKI) 设计过程中至关重要。在这项研究中,我们提出了一种从晶体结构中提取片段和氨基酸残基以进行激酶-配体相互作用的方法。此外,还提取了与激酶重要铰链区相互作用的核心片段及其装饰物。基于来自激酶-配体相互作用指纹和结构数据库的激酶的叠加结构数据,我们获得了两个文库,即包含 6672 对片段和相应氨基-的铰链非聚焦片段-氨基酸对文库 (FAP Lib)。酸,和一个以铰链为中心的铰链活页夹库(HB Lib),包含 3560 对铰链绑定支架及其相应的装饰。这两个库构成了一个以激酶为中心的相互作用数据库 (KID)。对 KID 进行了深入分析,以探索片段在 SMKI 设计中的重要特征。借助 KID,我们构建了两个以激酶为中心的分子数据库,一个名为 Recomb_DB,其中包含基于 FAP Lib 通过片段重组生成的 1,72,346 个分子,另一个名为 RsdHB_DB,其中包含基于我们的 HB Lib 使用分子生成生成的 93,030 个分子方法。与商业和非商业的五个数据库相比,这两个数据库在支架多样性方面均排名前三,在分子指纹多样性方面均排名前四,并且更侧重于激酶抑制剂的化学空间。











































 京公网安备 11010802027423号
京公网安备 11010802027423号