当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
System truncation accelerates binding affinity calculations with the fragment molecular orbital method: A benchmark study
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2022-11-29 , DOI: 10.1002/jcc.27044 Shinya Nakamura 1 , Tatsuo Akaki 1, 2 , Keiji Nishiwaki 1 , Midori Nakatani 1 , Yuji Kawase 1 , Yuki Takahashi 1 , Isao Nakanishi 1
Journal of Computational Chemistry ( IF 3 ) Pub Date : 2022-11-29 , DOI: 10.1002/jcc.27044 Shinya Nakamura 1 , Tatsuo Akaki 1, 2 , Keiji Nishiwaki 1 , Midori Nakatani 1 , Yuji Kawase 1 , Yuki Takahashi 1 , Isao Nakanishi 1
Affiliation
|
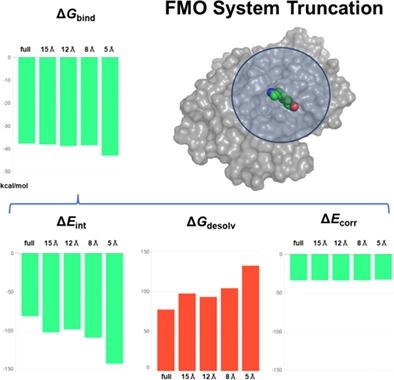
The fragment molecular orbital (FMO) method is a fast quantum-mechanical method that divides systems into pieces of fragments and performs ab initio calculations. The system truncation enables further speed improvement. In this article, we systematically study the effects of system truncations on binding affinity calculations obtained with FMO in combination with either the polarizable continuum model (FMO/PCM) or in combination with the Møller–Plesset method (FMO-MP2). We have used five protein complexes with ligands of several charged states. The calculated binding energies of the size variants of the truncated system, including only a restricted number of atoms around the ligand, are compared to the energy obtained from a full system. The result shows that the systems could be truncated to a radius of 8 Å from neutral ligands within an error of 0.7 kcal/mol, and 12 Å from charged ligands within an error of 1.1 kcal/mol for calculating the binding energy in solution.
中文翻译:

系统截断加速了片段分子轨道方法的结合亲和力计算:一项基准研究
片段分子轨道 (FMO) 方法是一种快速的量子力学方法,它将系统划分为片段片段并执行从头算计算。系统截断可以进一步提高速度。在这篇文章中,我们系统地研究了系统截断对结合亲和力计算的影响,结合亲和力计算是通过 FMO 结合可极化连续介质模型 (FMO/PCM) 或结合 Møller–Plesset 方法 (FMO-MP2) 获得的。我们使用了五种蛋白质复合物和几种带电状态的配体。将截断系统的大小变体(仅包括配体周围有限数量的原子)的计算结合能与从完整系统获得的能量进行比较。
更新日期:2022-11-29
中文翻译:

系统截断加速了片段分子轨道方法的结合亲和力计算:一项基准研究
片段分子轨道 (FMO) 方法是一种快速的量子力学方法,它将系统划分为片段片段并执行从头算计算。系统截断可以进一步提高速度。在这篇文章中,我们系统地研究了系统截断对结合亲和力计算的影响,结合亲和力计算是通过 FMO 结合可极化连续介质模型 (FMO/PCM) 或结合 Møller–Plesset 方法 (FMO-MP2) 获得的。我们使用了五种蛋白质复合物和几种带电状态的配体。将截断系统的大小变体(仅包括配体周围有限数量的原子)的计算结合能与从完整系统获得的能量进行比较。



























 京公网安备 11010802027423号
京公网安备 11010802027423号