当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Decoding the Identification Mechanism of an SAM-III Riboswitch on Ligands through Multiple Independent Gaussian-Accelerated Molecular Dynamics Simulations
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-28 , DOI: 10.1021/acs.jcim.2c00961 Jianzhong Chen 1 , Qingkai Zeng 1 , Wei Wang 1 , Haibo Sun 1 , Guodong Hu 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-28 , DOI: 10.1021/acs.jcim.2c00961 Jianzhong Chen 1 , Qingkai Zeng 1 , Wei Wang 1 , Haibo Sun 1 , Guodong Hu 2
Affiliation
|
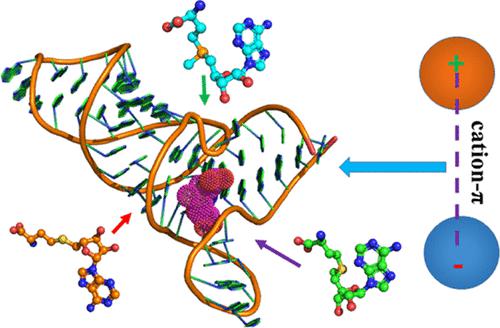
S-Adenosyl-l-methionine (SAM)-responsive riboswitches play a central role in the regulation of bacterial gene expression at the level of transcription attenuation or translation inhibition. In this study, multiple independent Gaussian-accelerated molecular dynamics simulations were performed to decipher the identification mechanisms of SAM-III (SMK) on ligands SAM, SAH, and EEM. The results reveal that ligand binding highly affects the structural flexibility, internal dynamics, and conformational changes of SAM-III. The dynamic analysis shows that helices P3 and P4 as well as two junctions J23 and J24 of SAM-III are highly susceptible to ligand binding. Analyses of free energy landscapes suggest that ligand binding induces different free energy profiles of SAM-III, which leads to the difference in identification sites of SAM-III on ligands. The information on ligand–nucleotide interactions not only uncovers that the π–π, cation−π, and hydrogen bonding interactions drive identification of SAM-III on the three ligands but also reveals that different electrostatic properties of SAM, SAH, and EEM alter the active sites of SAM-III. Meanwhile, the results also verify that the adenine group of SAM, SAH, and EEM is well recognized by conserved nucleotides G7, A29, U37, A38, and G48. We expect that this study can provide useful information for understanding the applications of SAM-III in chemical, synthetic RNA biology, and biomedical fields.
中文翻译:

通过多个独立的高斯加速分子动力学模拟解码配体上 SAM-III 核糖开关的识别机制
S-腺苷-l-甲硫氨酸 (SAM) 反应性核糖开关在转录减弱或翻译抑制水平的细菌基因表达调控中起着核心作用。在这项研究中,进行了多个独立的高斯加速分子动力学模拟,以破译 SAM-III (S MK ) 的识别机制) 在配体 SAM、SAH 和 EEM 上。结果表明,配体结合高度影响 SAM-III 的结构灵活性、内部动力学和构象变化。动态分析表明,螺旋 P3 和 P4 以及 SAM-III 的两个连接点 J23 和 J24 对配体结合高度敏感。自由能景观分析表明,配体结合会导致 SAM-III 的不同自由能分布,从而导致 SAM-III 在配体上的识别位点不同。有关配体-核苷酸相互作用的信息不仅揭示了 π-π、阳离子-π 和氢键相互作用驱动了三种配体上 SAM-III 的识别,而且揭示了 SAM、SAH 和 EEM 的不同静电特性改变了SAM-III 的活性位点。同时,该结果也验证了SAM的腺嘌呤基团,SAH 和 EEM 被保守核苷酸 G7、A29、U37、A38 和 G48 很好地识别。我们期望这项研究能够为理解 SAM-III 在化学、合成 RNA 生物学和生物医学领域的应用提供有用的信息。
更新日期:2022-11-28
中文翻译:

通过多个独立的高斯加速分子动力学模拟解码配体上 SAM-III 核糖开关的识别机制
S-腺苷-l-甲硫氨酸 (SAM) 反应性核糖开关在转录减弱或翻译抑制水平的细菌基因表达调控中起着核心作用。在这项研究中,进行了多个独立的高斯加速分子动力学模拟,以破译 SAM-III (S MK ) 的识别机制) 在配体 SAM、SAH 和 EEM 上。结果表明,配体结合高度影响 SAM-III 的结构灵活性、内部动力学和构象变化。动态分析表明,螺旋 P3 和 P4 以及 SAM-III 的两个连接点 J23 和 J24 对配体结合高度敏感。自由能景观分析表明,配体结合会导致 SAM-III 的不同自由能分布,从而导致 SAM-III 在配体上的识别位点不同。有关配体-核苷酸相互作用的信息不仅揭示了 π-π、阳离子-π 和氢键相互作用驱动了三种配体上 SAM-III 的识别,而且揭示了 SAM、SAH 和 EEM 的不同静电特性改变了SAM-III 的活性位点。同时,该结果也验证了SAM的腺嘌呤基团,SAH 和 EEM 被保守核苷酸 G7、A29、U37、A38 和 G48 很好地识别。我们期望这项研究能够为理解 SAM-III 在化学、合成 RNA 生物学和生物医学领域的应用提供有用的信息。










































 京公网安备 11010802027423号
京公网安备 11010802027423号