Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2022-11-21 , DOI: 10.1016/j.aca.2022.340636 Xun Liao 1 , Xiaolin Bai 1 , Shuguan Wang 2 , Christany Liggins 2 , Li Pan 2 , Meiyuan Wang 2 , Paul Tchounwou 3 , Jinghe Mao 4 , Yi-Ming Liu 2
|
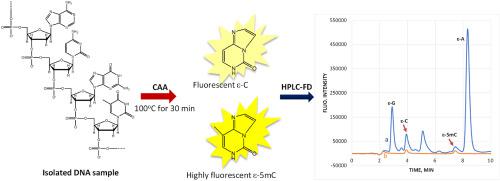
DNA methylation is intensively studied in medical science. Current HPLC methods for quantification of global DNA methylation involve digestion of a DNA sample and HPLC determination of both cytosine (C) and 5-methylcytosine (5mC) so that percentage of 5mC in total cytosine can be calculated as DNA methylation level. Herein we report a novel HPLC method based on a one-pot fluorescence tagging and depyrimidination reaction between DNA and chloroacetaldehyde (CAA) for highly sensitive quantification of global DNA methylation. In the one-pot reaction, C and 5mC residues in a DNA sequence react with CAA, forming fluorescent etheno-adducts that are then released from the sequence through depyrimidination. Interestingly, etheno-5mC (ε-5mC) is ∼20 times more fluorescent than ε-C and other ε-nucleobases resulting from the reaction, which greatly facilitates the quantification. Further, due to the tagging-induced increase in structural aromaticity, ε-nucleobases are far more separable by HPLC than intact nucleobases. The proposed HPLC method with fluorescence detection (HPLC-FD) is quick (i.e., < 1h per assay) and highly sensitive with a detection limit of 0.80 nM (or 250 fg on column) for 5mC. Using the method, DNA samples isolated from yeast, HCT-116 cells, and tissues were analyzed. Global DNA methylation was measured to be in the range from 0.35% to 2.23% in the samples analyzed. This sensitive method allowed accurate analyses of minute DNA samples (∼100 ng) isolated from milligrams of tissues.
中文翻译:

一种新颖的一锅荧光标记和去嘧啶策略,用于定量整体 DNA 甲基化
DNA 甲基化在医学领域得到深入研究。目前用于定量整体 DNA 甲基化的 HPLC 方法涉及 DNA 样品的消化和胞嘧啶 (C) 和 5-甲基胞嘧啶 (5mC) 的 HPLC 测定,以便可以计算总胞嘧啶中 5mC 的百分比作为 DNA 甲基化水平。在此,我们报告了一种基于 DNA 和氯乙醛 (CAA) 之间的一锅荧光标记和脱嘧啶反应的新型 HPLC 方法,用于高灵敏度地定量整体 DNA 甲基化。在一锅反应中,DNA 序列中的 C 和 5mC 残基与 CAA 反应,形成荧光乙烯加合物,然后通过脱嘧啶作用从序列中释放出来。有趣的是,etheno-5mC (ε-5mC) 的荧光比反应产生的 ε-C 和其他 ε-核碱基强约 20 倍,这极大地促进了定量。此外,由于标记诱导的结构芳香性增加,ε-核碱基通过 HPLC 比完整核碱基更容易分离。所提出的荧光检测 HPLC 方法 (HPLC-FD) 速度快(即每次测定 < 1 小时)且灵敏度高,5 mC 的检测限为 0.80 nM(或柱上 250 fg)。使用该方法,对从酵母、HCT-116 细胞和组织中分离的 DNA 样本进行了分析。在分析的样品中,整体 DNA 甲基化程度被测量为 0.35% 至 2.23%。这种灵敏的方法可以准确分析从毫克组织中分离出的微小 DNA 样本(∼100 ng)。









































 京公网安备 11010802027423号
京公网安备 11010802027423号