当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
RGN: Residue-Based Graph Attention and Convolutional Network for Protein–Protein Interaction Site Prediction
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-18 , DOI: 10.1021/acs.jcim.2c01092 Shuang Wang 1 , Wenqi Chen 1 , Peifu Han 1 , Xue Li 1 , Tao Song 1, 2
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-18 , DOI: 10.1021/acs.jcim.2c01092 Shuang Wang 1 , Wenqi Chen 1 , Peifu Han 1 , Xue Li 1 , Tao Song 1, 2
Affiliation
|
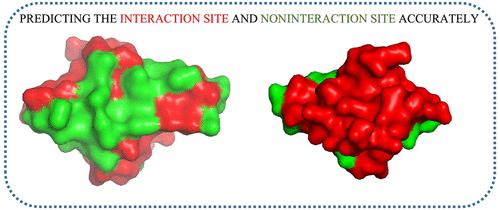
The prediction of a protein–protein interaction site (PPI site) plays a very important role in the biochemical process, and lots of computational methods have been proposed in the past. However, the majority of the past methods are time consuming and lack accuracy. Hence, coming up with an effective computational method is necessary. In this article, we present a novel computational model called RGN (residue-based graph attention and convolutional network) to predict PPI sites. In our paper, the protein is treated as a graph. The amino acid can be seen as the node in the graph structure. The position-specific scoring matrix, hidden Markov model, hydrogen bond estimation algorithm, and ProtBert are applied as node features. The edges are decided by the spatial distance between the amino acids. Then, we utilize a residue-based graph convolutional network and graph attention network to further extract the deeper feature. Finally, the processed node feature is fed into the prediction layer. We show the superiority of our model by comparing it with the other four protein structure-based methods and five protein sequence-based methods. Our model obtains the best performance on all the evaluation metrics (accuracy, precision, recall, F1 score, Matthews correlation coefficient, area under the receiver operating characteristic curve, and area under the precision recall curve). We also conduct a case study to demonstrate that extracting the protein information from the protein structure perspective is effective and points out the difficult aspect of PPI site prediction.
中文翻译:

RGN:用于蛋白质-蛋白质相互作用位点预测的基于残基的图注意力和卷积网络
蛋白质-蛋白质相互作用位点(PPI位点)的预测在生化过程中起着非常重要的作用,过去已经提出了许多计算方法。然而,过去的大多数方法都非常耗时且缺乏准确性。因此,有必要提出一种有效的计算方法。在本文中,我们提出了一种称为 RGN(基于残差的图注意力和卷积网络)的新型计算模型来预测 PPI 位点。在我们的论文中,蛋白质被视为图形。氨基酸可以看作是图结构中的节点。特定位置的评分矩阵、隐马尔可夫模型、氢键估计算法和 ProtBert 被用作节点特征。边缘由氨基酸之间的空间距离决定。然后,我们利用基于残差的图卷积网络和图注意力网络来进一步提取更深层次的特征。最后,处理后的节点特征被送入预测层。通过与其他四种基于蛋白质结构的方法和五种基于蛋白质序列的方法进行比较,我们展示了我们模型的优越性。我们的模型在所有评估指标(准确性、精确度、召回率、F 1得分、Matthews 相关系数、接受者操作特征曲线下的面积和精确召回曲线下的面积)。我们还进行了一个案例研究,以证明从蛋白质结构的角度提取蛋白质信息是有效的,并指出了 PPI 位点预测的难点。
更新日期:2022-11-18
中文翻译:

RGN:用于蛋白质-蛋白质相互作用位点预测的基于残基的图注意力和卷积网络
蛋白质-蛋白质相互作用位点(PPI位点)的预测在生化过程中起着非常重要的作用,过去已经提出了许多计算方法。然而,过去的大多数方法都非常耗时且缺乏准确性。因此,有必要提出一种有效的计算方法。在本文中,我们提出了一种称为 RGN(基于残差的图注意力和卷积网络)的新型计算模型来预测 PPI 位点。在我们的论文中,蛋白质被视为图形。氨基酸可以看作是图结构中的节点。特定位置的评分矩阵、隐马尔可夫模型、氢键估计算法和 ProtBert 被用作节点特征。边缘由氨基酸之间的空间距离决定。然后,我们利用基于残差的图卷积网络和图注意力网络来进一步提取更深层次的特征。最后,处理后的节点特征被送入预测层。通过与其他四种基于蛋白质结构的方法和五种基于蛋白质序列的方法进行比较,我们展示了我们模型的优越性。我们的模型在所有评估指标(准确性、精确度、召回率、F 1得分、Matthews 相关系数、接受者操作特征曲线下的面积和精确召回曲线下的面积)。我们还进行了一个案例研究,以证明从蛋白质结构的角度提取蛋白质信息是有效的,并指出了 PPI 位点预测的难点。











































 京公网安备 11010802027423号
京公网安备 11010802027423号