Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2022-11-12 , DOI: 10.1016/j.csbj.2022.11.019 Alexandre Smirnov 1, 2
|
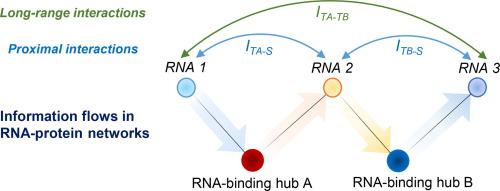
RNA-binding proteins are central players in post-transcriptional regulation. Some of them, such as the well-studied bacterial RNA chaperones Hfq and ProQ or the eukaryotic RNAi factor Argonaute, interact with hundreds-to-thousands of different RNAs and thereby globally affect gene expression. As a shared yet limited resource, these and other RNA-binding hubs drive strong competition between their multiple ligands. This creates a ground for significant cross-communication between RNA targets, which enables them to share information, “synchronise” their behaviour, and produce interesting biochemical effects, sometimes propagating across the highly connected RNA-protein network. This property is likely universally present in hub-centred networks and plays a key role in global gene expression programmes. It is also an important factor in biotechnology and synthetic biology applications of RNA/protein-based circuits. However, few studies so-far focused on describing and explaining this phenomenon from first principles. Here we introduce an information theory-based framework to comprehensively and exactly describe information flow in hub-centred networks. We show that information sharing can achieve significant levels in relatively small networks, provided the hub is present in limiting concentrations. The transmitted information is sufficient to noticeably affect the binding probabilities of competing targets but drops exponentially along the network. Target overexpression can disrupt communication between other targets, while hub sequestration boosts the crosstalk. We also find that overlaps between the interactomes of two different hubs create both entropic challenges and new forms of long-range communication between RNAs and proteins.
中文翻译:

全局 RNA 结合蛋白如何协调 RNA 调节子的行为:一种信息方法
RNA 结合蛋白是转录后调控的核心参与者。其中一些,例如研究充分的细菌 RNA 伴侣 Hfq 和 ProQ 或真核 RNAi 因子 Argonaute,与成百上千种不同的 RNA 相互作用,从而在全球范围内影响基因表达。作为一种共享但有限的资源,这些和其他 RNA 结合中心推动了它们的多个配体之间的激烈竞争。这为 RNA 目标之间的重要交叉通信创造了基础,使它们能够共享信息、“同步”它们的行为并产生有趣的生化效应,有时会在高度连接的 RNA-蛋白质网络中传播。此属性可能普遍存在于以枢纽为中心的网络中,并在全球基因表达计划中发挥关键作用。它也是基于 RNA/蛋白质的电路的生物技术和合成生物学应用中的一个重要因素。然而,迄今为止,很少有研究着眼于从第一性原理来描述和解释这一现象。在这里,我们引入了一个基于信息论的框架来全面准确地描述以枢纽为中心的网络中的信息流。我们表明,信息共享可以在相对较小的网络中达到显着水平,前提是枢纽以有限的浓度存在。传输的信息足以显着影响竞争目标的结合概率,但会沿网络呈指数下降。目标过度表达会破坏其他目标之间的通信,而中心隔离会增加串扰。











































 京公网安备 11010802027423号
京公网安备 11010802027423号