Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
How an ACE2 mimicking epitope-MIP nanofilm recognizes template-related peptides and the receptor binding domain of SARS-CoV-2
Nanoscale ( IF 5.8 ) Pub Date : 2022-11-15 , DOI: 10.1039/d2nr03898f Xiaorong Zhang 1 , Armel T Waffo 2 , Aysu Yarman 1, 3 , Norbert Kovács 4 , Zsófia Bognár 4, 5 , Ulla Wollenberger 1 , Ibrahim M El-Sherbiny 6 , Rabeay Y A Hassan 6 , Frank F Bier 1 , Róbert E Gyurcsányi 4, 5 , Ingo Zebger 2 , Frieder W Scheller 1
Nanoscale ( IF 5.8 ) Pub Date : 2022-11-15 , DOI: 10.1039/d2nr03898f Xiaorong Zhang 1 , Armel T Waffo 2 , Aysu Yarman 1, 3 , Norbert Kovács 4 , Zsófia Bognár 4, 5 , Ulla Wollenberger 1 , Ibrahim M El-Sherbiny 6 , Rabeay Y A Hassan 6 , Frank F Bier 1 , Róbert E Gyurcsányi 4, 5 , Ingo Zebger 2 , Frieder W Scheller 1
Affiliation
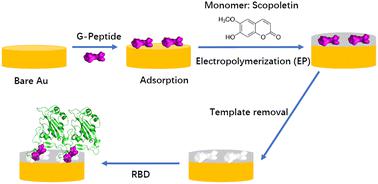
|
Here we aim to gain a mechanistic understanding of the formation of epitope-imprinted polymer nanofilms using a non-terminal peptide sequence, i.e. the peptide GFNCYFP (G485 to P491) of the SARS-CoV-2 receptor binding domain (RBD). This epitope is chemisorbed on the gold surface through the central cysteine 488 followed by the electrosynthesis of a ∼5 nm thick polyscopoletin film around the surface confined templates. The interaction of peptides and the parent RBD and spike protein with the imprinted polyscopoletin nanofilm was followed by electrochemical redox marker gating, surface enhanced infrared absorption spectroscopy and conductive AFM. Because the use of non-terminal epitopes is especially intricate, here we characterize the binding pockets through their interaction with 5 peptides rationally derived from the template sequence, i.e. implementing central single amino acid mismatch as well as elongations and truncations at its C- and N- termini. Already a single amino acid mismatch, i.e. the central Cys488 substituted by a serine, results in ca. 15-fold lower affinity. Further truncation of the peptides to tetrapeptide (EGFN) and hexapeptide (YFPLQS) results also in a significantly lower affinity. We concluded that the affinity towards the different peptides is mainly determined by the four amino acid motif CYFP present in the sequence of the template peptide. A higher affinity than that for the peptides is found for the parent proteins RBD and spike protein, which seems to be due to out of cavity effects caused by their larger footprint on the nanofilm surface.
中文翻译:

ACE2 模拟表位-MIP 纳米膜如何识别模板相关肽和 SARS-CoV-2 的受体结合域
在这里,我们旨在获得对使用非末端肽序列形成表位印迹聚合物纳米膜的机制的理解,即SARS-CoV-2 受体结合域 (RBD) 的肽 GFNCYFP(G485 至 P491)。该表位通过中央半胱氨酸 488 化学吸附在金表面上,然后在表面限制模板周围电合成约 5 nm 厚的聚东莨菪碱膜。通过电化学氧化还原标记门控、表面增强红外吸收光谱和导电 AFM,跟踪肽与母体 RBD 和刺突蛋白与印迹聚东莨菪碱纳米膜的相互作用。因为非末端表位的使用特别复杂,我们在这里通过它们与从模板序列合理衍生的 5 个肽的相互作用来表征结合口袋,即在其 C- 和 N- 末端实施中央单个氨基酸错配以及延伸和截断。已经有一个氨基酸错配,即中心 Cys488 被丝氨酸取代,导致ca。亲和力降低 15 倍。将肽进一步截短为四肽 (EGFN) 和六肽 (YFPLQS) 也会导致亲和力显着降低。我们得出结论,对不同肽的亲和力主要由模板肽序列中存在的四个氨基酸基序 CYFP 决定。发现母体蛋白 RBD 和刺突蛋白的亲和力高于肽,这似乎是由于它们在纳米膜表面上的足迹较大而导致的腔外效应。
更新日期:2022-11-15
中文翻译:

ACE2 模拟表位-MIP 纳米膜如何识别模板相关肽和 SARS-CoV-2 的受体结合域
在这里,我们旨在获得对使用非末端肽序列形成表位印迹聚合物纳米膜的机制的理解,即SARS-CoV-2 受体结合域 (RBD) 的肽 GFNCYFP(G485 至 P491)。该表位通过中央半胱氨酸 488 化学吸附在金表面上,然后在表面限制模板周围电合成约 5 nm 厚的聚东莨菪碱膜。通过电化学氧化还原标记门控、表面增强红外吸收光谱和导电 AFM,跟踪肽与母体 RBD 和刺突蛋白与印迹聚东莨菪碱纳米膜的相互作用。因为非末端表位的使用特别复杂,我们在这里通过它们与从模板序列合理衍生的 5 个肽的相互作用来表征结合口袋,即在其 C- 和 N- 末端实施中央单个氨基酸错配以及延伸和截断。已经有一个氨基酸错配,即中心 Cys488 被丝氨酸取代,导致ca。亲和力降低 15 倍。将肽进一步截短为四肽 (EGFN) 和六肽 (YFPLQS) 也会导致亲和力显着降低。我们得出结论,对不同肽的亲和力主要由模板肽序列中存在的四个氨基酸基序 CYFP 决定。发现母体蛋白 RBD 和刺突蛋白的亲和力高于肽,这似乎是由于它们在纳米膜表面上的足迹较大而导致的腔外效应。











































 京公网安备 11010802027423号
京公网安备 11010802027423号