当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
An Integrated Machine Learning Model To Spot Peptide Binding Pockets in 3D Protein Screening
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-01 , DOI: 10.1021/acs.jcim.2c00583 Daniela Trisciuzzi 1, 2 , Lydia Siragusa 2, 3 , Massimo Baroni 2 , Gabriele Cruciani 4 , Orazio Nicolotti 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-11-01 , DOI: 10.1021/acs.jcim.2c00583 Daniela Trisciuzzi 1, 2 , Lydia Siragusa 2, 3 , Massimo Baroni 2 , Gabriele Cruciani 4 , Orazio Nicolotti 1
Affiliation
|
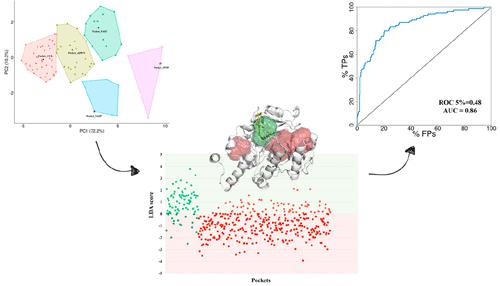
The prediction of peptide–protein binding sites is of utmost importance to tackle the onset of severe neurodegenerative diseases and cancer. In this work, we detail a novel machine learning model based on Linear Discriminant Analysis (LDA) demonstrating to be highly predictive in detecting the putative protein binding regions of small peptides. Starting from 439 high-quality pockets derived from peptide–protein crystallographic complexes, three sets of well-established peptide-binding regions were first selected through a Partitioning Around Medoids (PAM) clustering algorithm based on morphological and energetic 3D GRID-MIF molecular descriptors. Next, the best combination between all the putative interacting peptide pockets and related GRID-MIF scores was automatically explored by using the LDA-based protocol implemented in BioGPS. This approach proved successful to recognize the actual interacting peptide regions (that is, AUC = 0.86 and partial ROC enrichment at 5% of 0.48) from all the other pockets of the protein. Validated on two external collections sets, including 445 and 347 crystallographic peptide–protein complexes, our LDA-based model could be effective to further run peptide–protein virtual screening campaigns.
中文翻译:

在 3D 蛋白质筛选中发现肽结合袋的集成机器学习模型
肽-蛋白质结合位点的预测对于解决严重神经退行性疾病和癌症的发作至关重要。在这项工作中,我们详细介绍了一种基于线性判别分析 (LDA) 的新型机器学习模型,该模型证明在检测小肽的假定蛋白质结合区域方面具有高度预测性。从源自肽-蛋白质晶体复合物的 439 个高质量口袋开始,首先通过基于形态学和能量 3D GRID-MIF 分子描述符的围绕中心点 (PAM) 聚类算法选择了三组完善的肽结合区域。接下来,通过使用在 BioGPS 中实施的基于 LDA 的协议,自动探索所有假定的相互作用肽袋和相关 GRID-MIF 分数之间的最佳组合。这种方法被证明可以成功地从蛋白质的所有其他口袋中识别实际相互作用的肽区域(即 AUC = 0.86 和部分 ROC 富集在 0.48 的 5%)。我们基于 LDA 的模型在两个外部集合集(包括 445 和 347 晶体肽-蛋白质复合物)上得到验证,可以有效地进一步运行肽-蛋白质虚拟筛选活动。
更新日期:2022-11-01
中文翻译:

在 3D 蛋白质筛选中发现肽结合袋的集成机器学习模型
肽-蛋白质结合位点的预测对于解决严重神经退行性疾病和癌症的发作至关重要。在这项工作中,我们详细介绍了一种基于线性判别分析 (LDA) 的新型机器学习模型,该模型证明在检测小肽的假定蛋白质结合区域方面具有高度预测性。从源自肽-蛋白质晶体复合物的 439 个高质量口袋开始,首先通过基于形态学和能量 3D GRID-MIF 分子描述符的围绕中心点 (PAM) 聚类算法选择了三组完善的肽结合区域。接下来,通过使用在 BioGPS 中实施的基于 LDA 的协议,自动探索所有假定的相互作用肽袋和相关 GRID-MIF 分数之间的最佳组合。这种方法被证明可以成功地从蛋白质的所有其他口袋中识别实际相互作用的肽区域(即 AUC = 0.86 和部分 ROC 富集在 0.48 的 5%)。我们基于 LDA 的模型在两个外部集合集(包括 445 和 347 晶体肽-蛋白质复合物)上得到验证,可以有效地进一步运行肽-蛋白质虚拟筛选活动。











































 京公网安备 11010802027423号
京公网安备 11010802027423号