当前位置:
X-MOL 学术
›
J. Integr. Plant Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A prolific and robust whole-genome genotyping method using PCR amplification via primer-template mismatched annealing
Journal of Integrative Plant Biology ( IF 9.3 ) Pub Date : 2022-10-21 , DOI: 10.1111/jipb.13395 Sheng Zhao 1, 2 , Cuicui Zhang 2 , Liqun Wang 1 , Minxuan Luo 1, 2 , Peng Zhang 2 , Yue Wang 2 , Waqar Afzal Malik 2 , Yue Wang 2 , Peng Chen 2 , Xianjin Qiu 3 , Chongrong Wang 4 , Hong Lu 2 , Yong Xiang 2 , Yuwen Liu 2 , Jue Ruan 2 , Qian Qian 2 , Haijian Zhi 1 , Yuxiao Chang 2
Journal of Integrative Plant Biology ( IF 9.3 ) Pub Date : 2022-10-21 , DOI: 10.1111/jipb.13395 Sheng Zhao 1, 2 , Cuicui Zhang 2 , Liqun Wang 1 , Minxuan Luo 1, 2 , Peng Zhang 2 , Yue Wang 2 , Waqar Afzal Malik 2 , Yue Wang 2 , Peng Chen 2 , Xianjin Qiu 3 , Chongrong Wang 4 , Hong Lu 2 , Yong Xiang 2 , Yuwen Liu 2 , Jue Ruan 2 , Qian Qian 2 , Haijian Zhi 1 , Yuxiao Chang 2
Affiliation
|
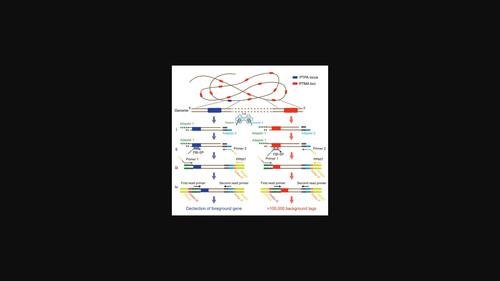
Whole-genome genotyping methods are important for breeding. However, it has been challenging to develop a robust method for simultaneous foreground and background genotyping that can easily be adapted to different genes and species. In our study, we accidently discovered that in adapter ligation-mediated PCR, the amplification by primer-template mismatched annealing (PTMA) along the genome could generate thousands of stable PCR products. Based on this observation, we consequently developed a novel method for simultaneous foreground and background integrated genotyping by sequencing (FBI-seq) using one specific primer, in which foreground genotyping is performed by primer-template perfect annealing (PTPA), while background genotyping employs PTMA. Unlike DNA arrays, multiple PCR, or genome target enrichments, FBI-seq requires little preliminary work for primer design and synthesis, and it is easily adaptable to different foreground genes and species. FBI-seq therefore provides a prolific, robust, and accurate method for simultaneous foreground and background genotyping to facilitate breeding in the post-genomics era.
中文翻译:

一种通过引物模板错配退火进行 PCR 扩增的多产且稳健的全基因组基因分型方法
全基因组基因分型方法对育种很重要。然而,开发一种可轻松适应不同基因和物种的同时前景和背景基因分型的稳健方法一直具有挑战性。在我们的研究中,我们意外地发现,在接头连接介导的 PCR 中,通过引物模板错配退火 (PTMA) 沿着基因组进行扩增可以产生数千种稳定的 PCR 产物。基于这一观察,我们因此开发了一种使用一个特异性引物通过测序(FBI-seq)同时进行前景和背景整合基因分型的新方法,其中前景基因分型通过引物模板完美退火(PTPA)进行,而背景基因分型采用聚甲基丙烯酸甲酯。与 DNA 阵列、多重 PCR 或基因组目标富集不同,FBI-seq 对引物设计和合成的前期工作很少,并且很容易适应不同的前景基因和物种。因此,FBI-seq 为同时进行前景和背景基因分型提供了一种多产、稳健且准确的方法,以促进后基因组学时代的育种。
更新日期:2022-10-21
中文翻译:

一种通过引物模板错配退火进行 PCR 扩增的多产且稳健的全基因组基因分型方法
全基因组基因分型方法对育种很重要。然而,开发一种可轻松适应不同基因和物种的同时前景和背景基因分型的稳健方法一直具有挑战性。在我们的研究中,我们意外地发现,在接头连接介导的 PCR 中,通过引物模板错配退火 (PTMA) 沿着基因组进行扩增可以产生数千种稳定的 PCR 产物。基于这一观察,我们因此开发了一种使用一个特异性引物通过测序(FBI-seq)同时进行前景和背景整合基因分型的新方法,其中前景基因分型通过引物模板完美退火(PTPA)进行,而背景基因分型采用聚甲基丙烯酸甲酯。与 DNA 阵列、多重 PCR 或基因组目标富集不同,FBI-seq 对引物设计和合成的前期工作很少,并且很容易适应不同的前景基因和物种。因此,FBI-seq 为同时进行前景和背景基因分型提供了一种多产、稳健且准确的方法,以促进后基因组学时代的育种。











































 京公网安备 11010802027423号
京公网安备 11010802027423号