Molecular Therapy ( IF 12.1 ) Pub Date : 2022-08-13 , DOI: 10.1016/j.ymthe.2022.08.008 Colin K W Lim 1 , Tristan X McCallister 1 , Christian Saporito-Magriña 1 , Garrett D McPheron 1 , Ramya Krishnan 1 , M Alejandra Zeballos C 1 , Jackson E Powell 1 , Lindsay V Clark 2 , Pablo Perez-Pinera 3 , Thomas Gaj 4
|
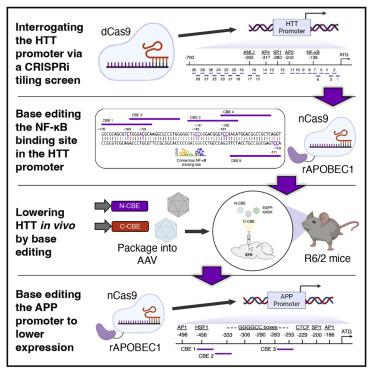
CRISPR technology has demonstrated broad utility for controlling target gene expression; however, there remains a need for strategies capable of modulating expression via the precise editing of non-coding regulatory elements. Here, we demonstrate that CRISPR base editors, a class of gene-modifying proteins capable of creating single-base substitutions in DNA, can be used to perturb gene expression via their targeted mutagenesis of cis-acting sequences. Using the promoter region of the human huntingtin (HTT) gene as an initial target, we show that editing of the binding site for the transcription factor NF-κB led to a marked reduction in HTT gene expression in base-edited cell populations. We found that these gene perturbations were persistent and specific, as a transcriptome-wide RNA analysis revealed minimal off-target effects resulting from the action of the base editor protein. We further demonstrate that this base-editing platform could influence gene expression in vivo as its delivery to a mouse model of Huntington’s disease led to a potent decrease in HTT mRNA in striatal neurons. Finally, to illustrate the applicability of this concept, we target the amyloid precursor protein, showing that multiplex editing of its promoter region significantly perturbed its expression. These findings demonstrate the potential for base editors to regulate target gene expression.
中文翻译:

顺式调控元件的 CRISPR 碱基编辑能够扰动神经变性相关基因
CRISPR 技术已在控制靶基因表达方面展现出广泛的用途;然而,仍然需要能够通过非编码调控元件的精确编辑来调节表达的策略。在这里,我们证明了 CRISPR 碱基编辑器(一类能够在 DNA 中创建单碱基替换的基因修饰蛋白)可用于通过顺式作用序列的定向诱变来扰乱基因表达。使用人类亨廷顿蛋白 (HTT) 基因的启动子区域作为初始靶标,我们发现转录因子 NF-κB 结合位点的编辑导致碱基编辑细胞群中 HTT 基因表达的显着降低。我们发现这些基因扰动是持久且特异的,因为全转录组 RNA 分析显示碱基编辑器蛋白的作用导致的脱靶效应最小。我们进一步证明,该碱基编辑平台可以影响体内基因表达,因为将其递送至亨廷顿病小鼠模型后,会导致纹状体神经元中 HTT mRNA 的有效减少。最后,为了说明这个概念的适用性,我们以淀粉样前体蛋白为目标,表明对其启动子区域的多重编辑显着扰乱了其表达。这些发现证明了碱基编辑器调节靶基因表达的潜力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号