当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A CRISPR/Cas12a-responsive dual-aptamer DNA network for specific capture and controllable release of circulating tumor cells
Chemical Science ( IF 7.6 ) Pub Date : 2022-08-12 , DOI: 10.1039/d2sc03374g Dong-Xia Wang 1 , Jing Wang 1, 2 , Ya-Xin Wang 1, 3 , Jia-Yi Ma 1 , Bo Liu 1 , An-Na Tang 1 , De-Ming Kong 1
Chemical Science ( IF 7.6 ) Pub Date : 2022-08-12 , DOI: 10.1039/d2sc03374g Dong-Xia Wang 1 , Jing Wang 1, 2 , Ya-Xin Wang 1, 3 , Jia-Yi Ma 1 , Bo Liu 1 , An-Na Tang 1 , De-Ming Kong 1
Affiliation
|
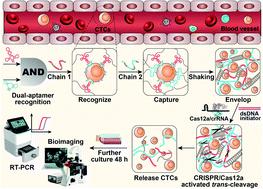
The separation and detection of circulating tumor cells (CTCs) have a significant impact on clinical diagnosis and treatment by providing a predictive diagnosis of primary tumors and tumor metastasis. But the responsive release and downstream analysis of live CTCs will provide more valuable information about molecular markers and functional properties. To this end, specific capture and controllable release methods, which can achieve the highly efficient enrichment of CTCs with strong viability, are urgently needed. DNA networks create a flexible, semi-wet three-dimensional (3D) microenvironment for cell culture, and have the potential to minimize the loss of cell viability and molecular integrity. More importantly, responsive DNA networks can be reasonably designed as smart sensors and devices to change shape, color, disassemble, and giving back to external stimuli. Here, a strategy for specifically collecting cells using a dual-aptamer DNA network is designed. The proposed strategy enables effective capture, 3D encapsulation, and responsive release of CTCs with strong viability, which can be used for downstream analysis of live cells. The programmability of CRISPR/Cas12a, a powerful toolbox for genome editing, is used to activate the responsive release of captured CTCs from the DNA network. After activation by a specified double-strand DNA (dsDNA) input, CRISPR/Cas12a cleaves the single-stranded DNA regions in the network, resulting in molecular to macroscopic changes in the network. Accompanied by the deconstruction of the DNA network into fragments, controllable cell release is achieved. The viability of released CTCs is well maintained and downstream cell analysis can be performed. This strategy uses the enzymatic properties of CRISPR/Cas12a to design a platform to improve the programmability and versatility of the DNA network, providing a powerful and effective method for capturing and releasing CTCs from complex physiological samples.
中文翻译:

用于特异性捕获和可控释放循环肿瘤细胞的 CRISPR/Cas12a 响应双适体 DNA 网络
循环肿瘤细胞(CTC)的分离和检测通过提供原发性肿瘤和肿瘤转移的预测性诊断,对临床诊断和治疗产生重大影响。但活 CTC 的响应释放和下游分析将提供有关分子标记和功能特性的更有价值的信息。为此,迫切需要特定的捕获和可控释放方法,以实现对具有强生存能力的CTCs的高效富集。DNA 网络为细胞培养创造了一个灵活的、半湿的三维 (3D) 微环境,并有可能最大限度地减少细胞活力和分子完整性的损失。更重要的是,响应式 DNA 网络可以合理地设计为智能传感器和设备,以改变形状、颜色、拆卸、并回馈外部刺激。在这里,设计了一种使用双适体 DNA 网络专门收集细胞的策略。所提出的策略能够有效捕获、3D 封装和响应释放具有强生存能力的 CTC,可用于活细胞的下游分析。CRISPR/Cas12a 是一个强大的基因组编辑工具箱,其可编程性用于激活从 DNA 网络中响应释放捕获的 CTC。在被指定的双链 DNA (dsDNA) 输入激活后,CRISPR/Cas12a 会切割网络中的单链 DNA 区域,从而导致网络发生分子到宏观的变化。伴随着DNA网络解构成片段,实现了可控的细胞释放。释放的 CTC 的活力得到很好的维持,可以进行下游细胞分析。该策略利用 CRISPR/Cas12a 的酶学特性设计了一个平台,以提高 DNA 网络的可编程性和多功能性,为从复杂生理样本中捕获和释放 CTC 提供了一种强大而有效的方法。
更新日期:2022-08-12
中文翻译:

用于特异性捕获和可控释放循环肿瘤细胞的 CRISPR/Cas12a 响应双适体 DNA 网络
循环肿瘤细胞(CTC)的分离和检测通过提供原发性肿瘤和肿瘤转移的预测性诊断,对临床诊断和治疗产生重大影响。但活 CTC 的响应释放和下游分析将提供有关分子标记和功能特性的更有价值的信息。为此,迫切需要特定的捕获和可控释放方法,以实现对具有强生存能力的CTCs的高效富集。DNA 网络为细胞培养创造了一个灵活的、半湿的三维 (3D) 微环境,并有可能最大限度地减少细胞活力和分子完整性的损失。更重要的是,响应式 DNA 网络可以合理地设计为智能传感器和设备,以改变形状、颜色、拆卸、并回馈外部刺激。在这里,设计了一种使用双适体 DNA 网络专门收集细胞的策略。所提出的策略能够有效捕获、3D 封装和响应释放具有强生存能力的 CTC,可用于活细胞的下游分析。CRISPR/Cas12a 是一个强大的基因组编辑工具箱,其可编程性用于激活从 DNA 网络中响应释放捕获的 CTC。在被指定的双链 DNA (dsDNA) 输入激活后,CRISPR/Cas12a 会切割网络中的单链 DNA 区域,从而导致网络发生分子到宏观的变化。伴随着DNA网络解构成片段,实现了可控的细胞释放。释放的 CTC 的活力得到很好的维持,可以进行下游细胞分析。该策略利用 CRISPR/Cas12a 的酶学特性设计了一个平台,以提高 DNA 网络的可编程性和多功能性,为从复杂生理样本中捕获和释放 CTC 提供了一种强大而有效的方法。











































 京公网安备 11010802027423号
京公网安备 11010802027423号