Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High-efficiency and high-fidelity ssDNA circularisation via the pairing of five 3′-terminal bases to assist LR-LAMP for the genotyping of single-nucleotide polymorphisms
Analyst ( IF 3.6 ) Pub Date : 2022-08-09 , DOI: 10.1039/d2an01042a Taiwen Li 1 , Huayan Zou 1 , Jing Zhang 1 , Haixia Ding 1 , Cheng Li 1 , Xiangru Chen 1 , Yunzhou Li 1 , Wenzhuo Feng 1 , Koji Kageyama 2
Analyst ( IF 3.6 ) Pub Date : 2022-08-09 , DOI: 10.1039/d2an01042a Taiwen Li 1 , Huayan Zou 1 , Jing Zhang 1 , Haixia Ding 1 , Cheng Li 1 , Xiangru Chen 1 , Yunzhou Li 1 , Wenzhuo Feng 1 , Koji Kageyama 2
Affiliation
|
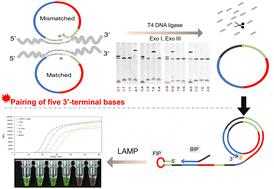
The poor fidelity of T4 DNA ligase has always limited the simple detection of single-nucleotide polymorphisms (SNPs) and is only applicable to some special SNP types. This study developed a highly sensitive and specific detection method for SNPs based on high-fidelity single-stranded circularisation. It used T4 DNA ligase and rolling circle amplification (RCA) plus loop-mediated isothermal amplification (LAMP). Surprisingly, the cyclisation stage's efficiency greatly improved. The ligation fidelity was almost perfect via the unique pairing pattern between a long-paired base at the 5′ terminus and only five bases at the 3′ terminus on linear single-stranded DNA (l-DNA). Subsequently, LR-LAMP was performed and combined with the circularisation step for the simple detection of SNPs. The results showed that even 100 aM targets could be detected correctly and that a mutation rate of 0.1% or even 0.01% could be analysed via naked-eye visualisation or fluorescence detection, respectively. In addition, genomic DNA samples were used to evaluate the method, which indicated that it could effectively distinguish the SNPs of RPA190-T1145A in Phytophthora infestans. This strategy may play an important role in both circularisation of single-stranded DNA and detecting arbitrary SNPs.
中文翻译:

通过五个 3'-末端碱基配对的高效和高保真 ssDNA 环化,以协助 LR-LAMP 进行单核苷酸多态性的基因分型
T4 DNA连接酶保真度差一直限制了单核苷酸多态性(SNPs)的简单检测,仅适用于一些特殊的SNP类型。本研究开发了一种基于高保真单链环化的 SNP 高灵敏度和特异性检测方法。它使用 T4 DNA 连接酶和滚环扩增 (RCA) 以及环介导的等温扩增 (LAMP)。令人惊讶的是,环化阶段的效率大大提高。连接保真度几乎是完美的线性单链 DNA (l-DNA) 上 5' 末端的长配对碱基与 3' 末端仅 5 个碱基之间的独特配对模式。随后,进行 LR-LAMP 并与环化步骤相结合,以简单检测 SNP。结果表明,即使是 100 个 aM 目标也可以被正确检测到,并且可以通过肉眼可视化或荧光检测分别分析 0.1% 甚至 0.01% 的突变率。此外,利用基因组DNA样本对该方法进行了评价,表明该方法可以有效区分致病疫霉中RPA190 -T1145A的SNP 。这种策略可能在单链 DNA 的环化和检测任意 SNP 中发挥重要作用。
更新日期:2022-08-09
中文翻译:

通过五个 3'-末端碱基配对的高效和高保真 ssDNA 环化,以协助 LR-LAMP 进行单核苷酸多态性的基因分型
T4 DNA连接酶保真度差一直限制了单核苷酸多态性(SNPs)的简单检测,仅适用于一些特殊的SNP类型。本研究开发了一种基于高保真单链环化的 SNP 高灵敏度和特异性检测方法。它使用 T4 DNA 连接酶和滚环扩增 (RCA) 以及环介导的等温扩增 (LAMP)。令人惊讶的是,环化阶段的效率大大提高。连接保真度几乎是完美的线性单链 DNA (l-DNA) 上 5' 末端的长配对碱基与 3' 末端仅 5 个碱基之间的独特配对模式。随后,进行 LR-LAMP 并与环化步骤相结合,以简单检测 SNP。结果表明,即使是 100 个 aM 目标也可以被正确检测到,并且可以通过肉眼可视化或荧光检测分别分析 0.1% 甚至 0.01% 的突变率。此外,利用基因组DNA样本对该方法进行了评价,表明该方法可以有效区分致病疫霉中RPA190 -T1145A的SNP 。这种策略可能在单链 DNA 的环化和检测任意 SNP 中发挥重要作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号