当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Systematic Analysis and Prediction of the Target Space of Bioactive Food Compounds: Filling the Chemobiological Gaps
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-08-08 , DOI: 10.1021/acs.jcim.2c00888 Andrés Sánchez-Ruiz 1 , Gonzalo Colmenarejo 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-08-08 , DOI: 10.1021/acs.jcim.2c00888 Andrés Sánchez-Ruiz 1 , Gonzalo Colmenarejo 1
Affiliation
|
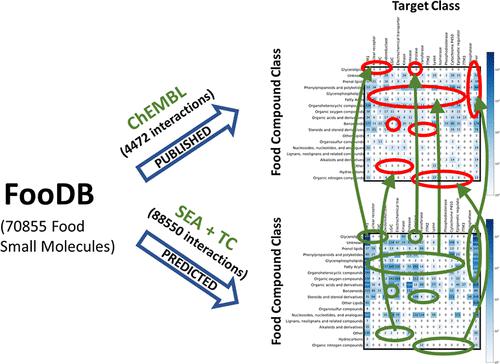
Food compounds and their molecular interactions are crucial for health and provide new chemotypes and targets for drug and nutraceutic design. Here, we retrieve and analyze the complete set of published interactions of food compounds with human proteins using the FooDB as a compound set and ChEMBL as a source of interactions. The data are analyzed in terms of 19 target classes and 19 compound classes, showing a small fraction of target assignment for the compounds (1.6%) and unraveling multiple gaps in the chemobiological space for these molecules. By using well-established cheminformatic approaches [similarity ensemble approach (SEA) combined with the maximum Tanimoto coefficient to the nearest bioactive, “SEA + TC”], we achieve a much enhanced target assignment (64.2%), filling many of the gaps with target hypothesis for fast focused testing. By publishing these data sets and analyses, we expect to provide a set of resources to speed up the full clarification of the chemobiological space of food compounds, opening new opportunities for drug and nutraceutic design.
中文翻译:

生物活性食品化合物目标空间的系统分析和预测:填补化学生物学空白
食品化合物及其分子相互作用对健康至关重要,并为药物和营养品设计提供新的化学类型和靶点。在这里,我们使用 FooDB 作为化合物集和 ChEMBL 作为相互作用的来源,检索和分析已发表的食物化合物与人类蛋白质的完整相互作用集。根据 19 个目标类别和 19 个化合物类别对数据进行了分析,显示了化合物的一小部分目标分配 (1.6%),并揭示了这些分子在化学生物学空间中的多个空白。通过使用成熟的化学信息学方法 [相似性集成方法 (SEA) 结合最接近的生物活性物质的最大 Tanimoto 系数,“SEA + TC”],我们实现了显着增强的目标分配 (64.2%),填补了许多空白快速集中测试的目标假设。
更新日期:2022-08-08
中文翻译:

生物活性食品化合物目标空间的系统分析和预测:填补化学生物学空白
食品化合物及其分子相互作用对健康至关重要,并为药物和营养品设计提供新的化学类型和靶点。在这里,我们使用 FooDB 作为化合物集和 ChEMBL 作为相互作用的来源,检索和分析已发表的食物化合物与人类蛋白质的完整相互作用集。根据 19 个目标类别和 19 个化合物类别对数据进行了分析,显示了化合物的一小部分目标分配 (1.6%),并揭示了这些分子在化学生物学空间中的多个空白。通过使用成熟的化学信息学方法 [相似性集成方法 (SEA) 结合最接近的生物活性物质的最大 Tanimoto 系数,“SEA + TC”],我们实现了显着增强的目标分配 (64.2%),填补了许多空白快速集中测试的目标假设。











































 京公网安备 11010802027423号
京公网安备 11010802027423号